SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells
- PMID: 16495264
- PMCID: PMC1426438
- DOI: 10.1128/AAC.50.3.1013-1020.2006
SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells
Abstract
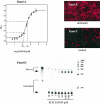
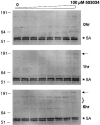
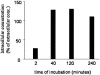
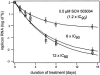
Cleavage of the hepatitis C virus (HCV) polyprotein by the viral NS3 protease releases functional viral proteins essential for viral replication. Recent studies by Foy and coworkers strongly suggest that NS3-mediated cleavage of host factors may abrogate cellular response to alpha interferon (IFN-alpha) (E. Foy, K. Li, R. Sumpter, Jr., Y.-M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr., Proc. Natl. Acad. Sci. USA 102:2986-2991, 2005, and E. Foy, K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr., Science 300:1145-1148, 2003). Blockage of NS3 protease activity therefore is expected to inhibit HCV replication by both direct suppression of viral protein production as well as by restoring host responsiveness to IFN. Using structure-assisted design, a ketoamide inhibitor, SCH 503034, was generated which demonstrated potent (overall inhibition constant, 14 nM) time-dependent inhibition of the NS3 protease in cell-free enzyme assays as well as robust in vitro activity in the HCV replicon system, as monitored by immunofluorescence and real-time PCR analysis. Continuous exposure of replicon-bearing cell lines to six times the 90% effective concentration of SCH 503034 for 15 days resulted in a greater than 4-log reduction in replicon RNA. The combination of SCH 503034 with IFN was more effective in suppressing replicon synthesis than either compound alone, supporting the suggestion of Foy and coworkers that combinations of IFN with protease inhibitors would lead to enhanced therapeutic efficacy.
Figures
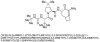


References
-
- Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. - PubMed
-
- Bain, V. G. 2001. Effect of HCV viral dynamics on treatment design: lessons learned from HIV. Am. J. Gastroenterol. 96:2818-2828. - PubMed
-
- Berger, A., and I. Schechter. 1970. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 257:249-264. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

