MicroRNAs direct rapid deadenylation of mRNA
- PMID: 16495412
- PMCID: PMC1449641
- DOI: 10.1073/pnas.0510928103
MicroRNAs direct rapid deadenylation of mRNA
Abstract
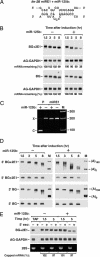
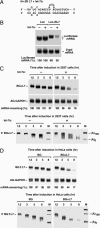
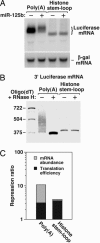
MicroRNAs (miRNAs) are ubiquitous regulators of eukaryotic gene expression. In addition to repressing translation, miRNAs can down-regulate the concentration of mRNAs that contain elements to which they are imperfectly complementary. Using miR-125b and let-7 as representative miRNAs, we show that in mammalian cells this reduction in message abundance is a consequence of accelerated deadenylation, which leads to rapid mRNA decay. The ability of miRNAs to expedite poly(A) removal does not result from decreased translation; nor does translational repression by miRNAs require a poly(A) tail, a 3' histone stem-loop being an effective substitute. These findings suggest that miRNAs use two distinct posttranscriptional mechanisms to down-regulate gene expression.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Comment in
-
MicroRNAs: new players in an old game.Proc Natl Acad Sci U S A. 2006 Mar 14;103(11):3951-2. doi: 10.1073/pnas.0601268103. Epub 2006 Mar 7. Proc Natl Acad Sci U S A. 2006. PMID: 16537465 Free PMC article. No abstract available.
References
-
- Zamore P. D., Tuschl T., Sharp P. A., Bartel D. P. Cell. 2000;101:25–33. - PubMed
-
- Lee R. C., Feinbaum R. L., Ambros V. Cell. 1993;75:843–854. - PubMed
-
- Wightman B., Ha I., Ruvkun G. Cell. 1993;75:855–862. - PubMed
-
- Moss E. G., Lee R. C., Ambros V. Cell. 1997;88:637–646. - PubMed
-
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Science. 2005;309:1573–1576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

