In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping
- PMID: 16498401
- PMCID: PMC1422155
- DOI: 10.1038/sj.emboj.7601021
In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping
Abstract
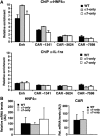
The gene encoding the nuclear receptor hepatocyte nuclear factor 4alpha (HNF4alpha) generates isoforms HNF4alpha1 and HNF4alpha7 from usage of alternative promoters. In particular, HNF4alpha7 is expressed in the pancreas whereas HNF4alpha1 is found in liver, and mutations affecting HNF4alpha function cause impaired insulin secretion and/or hepatic defects in humans and in tissue-specific 'knockout' mice. HNF4alpha1 and alpha7 isoforms differ exclusively by amino acids encoded by the first exon which, in HNF4alpha1 but not in HNF4alpha7, includes the activating function (AF)-1 transactivation domain. To investigate the roles of HNF4alpha1 and HNF4alpha7 in vivo, we generated mice expressing only one isoform under control of both promoters, via reciprocal swapping of the isoform-specific first exons. Unlike Hnf4alpha gene disruption which causes embryonic lethality, these 'alpha7-only' and 'alpha1-only' mice are viable, indicating functional redundancy of the isoforms. However, the former show dyslipidemia and preliminary results indicate impaired glucose tolerance for the latter, revealing functional specificities of the isoforms. These 'knock-in' mice provide the first test in vivo of the HNF4alpha AF-1 function and have permitted identification of AF-1-dependent target genes.
Figures




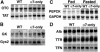

References
-
- Briancon N, Bailly A, Clotman F, Jacquemin P, Lemaigre FP, Weiss MC (2004) Expression of the α7 isoform of hepatocyte nuclear factor (HNF) 4 is activated by HNF6/OC-2 and HNF1 and repressed by HNF4α1 in the liver. J Biol Chem 279: 33398–33408 - PubMed
-
- Chabardes-Garonne D, Mejean A, Aude JC, Cheval L, Di Stefano A, Gaillard MC, Imbert-Teboul M, Wittner M, Balian C, Anthouard V, Robert C, Segurens B, Wincker P, Weissenbach J, Doucet A, Elalouf JM (2003) A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci USA 100: 13710–13715 - PMC - PubMed
-
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE Jr (1994) Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev 8: 2466–2477 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

