The gene encoding the fragile X RNA-binding protein is controlled by nuclear respiratory factor 2 and the CREB family of transcription factors
- PMID: 16500891
- PMCID: PMC1383620
- DOI: 10.1093/nar/gkj521
The gene encoding the fragile X RNA-binding protein is controlled by nuclear respiratory factor 2 and the CREB family of transcription factors
Abstract
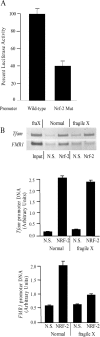
FMR1 encodes an RNA-binding protein whose absence results in fragile X mental retardation. In most patients, the FMR1 gene is cytosine-methylated and transcriptionally inactive. NRF-1 and Sp1 are known to bind and stimulate the active, but not the methylated/silenced, FMR1 promoter. Prior analysis has implicated a CRE site in regulation of FMR1 in neural cells but the role of this site is controversial. We now show that a phospho-CREB/ATF family member is bound to this site in vivo. We also find that the histone acetyltransferases CBP and p300 are associated with active FMR1 but are lost at the hypoacetylated fragile X allele. Surprisingly, FMR1 is not cAMP-inducible and resides in a newly recognized subclass of CREB-regulated genes. We have also elucidated a role for NRF-2 as a regulator of FMR1 in vivo through a previously unrecognized and highly conserved recognition site in FMR1. NRF-1 and NRF-2 act additively while NRF-2 synergizes with CREB/ATF at FMR1's promoter. These data add FMR1 to the collection of genes controlled by both NRF-1 and NRF-2 and disfavor its membership in the immediate early response group of genes.
Figures


References
-
- Willemsen R., Oostra B.A., Bassell G.J., Dictenberg J. The fragile X syndrome: from molecular genetics to neurobiology. Ment. Retard Dev. Disabil. Res. Rev. 2004;10:60–67. - PubMed
-
- Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. - PubMed
-
- Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;6:397–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

