Identification of Hepta- and Octo-Uridine stretches as sole signals for programmed +1 and -1 ribosomal frameshifting during translation of SARS-CoV ORF 3a variants
- PMID: 16500894
- PMCID: PMC1383626
- DOI: 10.1093/nar/gkl017
Identification of Hepta- and Octo-Uridine stretches as sole signals for programmed +1 and -1 ribosomal frameshifting during translation of SARS-CoV ORF 3a variants
Abstract
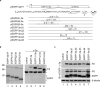
Programmed frameshifting is one of the translational recoding mechanisms that read the genetic code in alternative ways. This process is generally programmed by signals at defined locations in a specific mRNA. In this study, we report the identification of hepta- and octo-uridine stretches as sole signals for programmed +1 and -1 ribosomal frameshifting during translation of severe acute respiratory syndrome coronavirus (SARS-CoV) ORF 3a variants. SARS-CoV ORF 3a encodes a minor structural protein of 274 amino acids. Over the course of cloning and expression of the gene, a mixed population of clones with six, seven, eight and nine T stretches located 14 nt downstream of the initiation codon was found. In vitro and in vivo expression of clones with six, seven and eight Ts, respectively, showed the detection of the full-length 3a protein. Mutagenesis studies led to the identification of the hepta- and octo-uridine stretches as slippery sequences for efficient frameshifting. Interestingly, no stimulatory elements were found in the sequences upstream or downstream of the slippage site. When the hepta- and octo-uridine stretches were used to replace the original slippery sequence of the SARS-CoV ORF 1a and 1b, efficient frameshift events were observed. Furthermore, the efficiencies of frameshifting mediated by the hepta- and octo-uridine stretches were not affected by mutations introduced into a downstream stem-loop structure that totally abolish the frameshift event mediated by the original slippery sequence of ORF 1a and 1b. Taken together, this study identifies the hepta- and octo-uridine stretches that function as sole elements for efficient +1 and -1 ribosomal frameshift events.
Figures



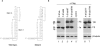

Similar articles
-
A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal.PLoS Biol. 2005 Jun;3(6):e172. doi: 10.1371/journal.pbio.0030172. Epub 2005 May 17. PLoS Biol. 2005. PMID: 15884978 Free PMC article.
-
A sequence required for -1 ribosomal frameshifting located four kilobases downstream of the frameshift site.J Mol Biol. 2001 Jul 27;310(5):987-99. doi: 10.1006/jmbi.2001.4801. J Mol Biol. 2001. PMID: 11502008
-
An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus.Nucleic Acids Res. 2005 Jul 29;33(13):4265-75. doi: 10.1093/nar/gki731. Print 2005. Nucleic Acids Res. 2005. PMID: 16055920 Free PMC article.
-
Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV.Virus Res. 2006 Jul;119(1):29-42. doi: 10.1016/j.virusres.2005.10.008. Epub 2005 Nov 28. Virus Res. 2006. PMID: 16310880 Free PMC article. Review.
-
The role of programmed-1 ribosomal frameshifting in coronavirus propagation.Front Biosci. 2008 May 1;13:4873-81. doi: 10.2741/3046. Front Biosci. 2008. PMID: 18508552 Free PMC article. Review.
Cited by
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
-
Binding of the 5'-untranslated region of coronavirus RNA to zinc finger CCHC-type and RNA-binding motif 1 enhances viral replication and transcription.Nucleic Acids Res. 2012 Jun;40(11):5065-77. doi: 10.1093/nar/gks165. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362731 Free PMC article.
-
Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus.PLoS Pathog. 2012;8(12):e1003065. doi: 10.1371/journal.ppat.1003065. Epub 2012 Dec 13. PLoS Pathog. 2012. PMID: 23271969 Free PMC article.
-
An infectious RNA with a hepta-adenosine stretch responsible for programmed -1 ribosomal frameshift derived from a full-length cDNA clone of Hibiscus latent Singapore virus.Virology. 2014 Jan 20;449:229-34. doi: 10.1016/j.virol.2013.11.021. Epub 2013 Dec 12. Virology. 2014. PMID: 24418557 Free PMC article.
-
The Emerging Roles of Viroporins in ER Stress Response and Autophagy Induction during Virus Infection.Viruses. 2015 Jun 4;7(6):2834-57. doi: 10.3390/v7062749. Viruses. 2015. PMID: 26053926 Free PMC article. Review.
References
-
- Craigen W.J., Caskey C.T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

