Sequence-dependent base pair opening in DNA double helix
- PMID: 16500982
- PMCID: PMC1432109
- DOI: 10.1529/biophysj.105.078774
Sequence-dependent base pair opening in DNA double helix
Abstract
Preservation of genetic information in DNA relies on shielding the nucleobases from damage within the double helix. Thermal fluctuations lead to infrequent events of the Watson-Crick basepair opening, or DNA "breathing", thus making normally buried groups available for modification and interaction with proteins. Fluctuational basepair opening implies the disruption of hydrogen bonds between the complementary bases and flipping of the base out of the helical stack. Prediction of sequence-dependent basepair opening probabilities in DNA is based on separation of the two major contributions to the stability of the double helix: lateral pairing between the complementary bases and stacking of the pairs along the helical axis. The partition function calculates the basepair opening probability at every position based on the loss of two stacking interactions and one base-pairing. Our model also includes a term accounting for the unfavorable positioning of the exposed base, which proceeds through a formation of a highly constrained small loop, or a ring. Quantitatively, the ring factor is found as an adjustable parameter from the comparison of the theoretical basepair opening probabilities and the experimental data on short DNA duplexes measured by NMR spectroscopy. We find that these thermodynamic parameters suggest nonobvious sequence dependent basepair opening probabilities.
Figures
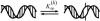

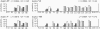

References
-
- Frank-Kamenetskii, M. D. 1987. How the double helix breathes. Nature. 328:17–18. - PubMed
-
- Frank-Kamenetskii, M. D. 1985. Fluctuational motility of DNA. In Structure and Motion: Membranes, Nucleic Acids and Proteins. E. Clementi, editor. Adenine Press, New York. 417–432.
-
- Gueron, M., M. Kochoyan, and J. L. Leroy. 1987. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 328:89–92. - PubMed
-
- Gueron, M., E. Charretier, J. Hagerhorst, M. Kochoyan, J. L. Leroy, and A. Moraillon. 1990. Applications of Imino Proton Exchange to Nucleic Acid Kinetics and Structures. In Structure and Methods. R. H. Sarma and M. H. Sarma, editors. Adenine Press, New York.: 113–137.
-
- Gueron, M., and J. L. Leroy. 1995. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 261:383–413. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

