Ion permeation through a narrow channel: using gramicidin to ascertain all-atom molecular dynamics potential of mean force methodology and biomolecular force fields
- PMID: 16500984
- PMCID: PMC1440729
- DOI: 10.1529/biophysj.105.077073
Ion permeation through a narrow channel: using gramicidin to ascertain all-atom molecular dynamics potential of mean force methodology and biomolecular force fields
Abstract
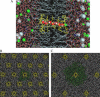
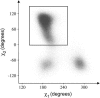
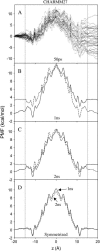
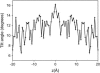
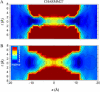
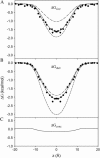
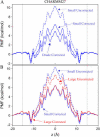
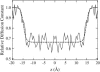
We investigate methods for extracting the potential of mean force (PMF) governing ion permeation from molecular dynamics simulations (MD) using gramicidin A as a prototypical narrow ion channel. It is possible to obtain well-converged meaningful PMFs using all-atom MD, which predict experimental observables within order-of-magnitude agreement with experimental results. This was possible by careful attention to issues of statistical convergence of the PMF, finite size effects, and lipid hydrocarbon chain polarizability. When comparing the modern all-atom force fields of CHARMM27 and AMBER94, we found that a fairly consistent picture emerges, and that both AMBER94 and CHARMM27 predict observables that are in semiquantitative agreement with both the experimental conductance and dissociation coefficient. Even small changes in the force field, however, result in significant changes in permeation energetics. Furthermore, the full two-dimensional free-energy surface describing permeation reveals the location and magnitude of the central barrier and the location of two binding sites for K(+) ion permeation near the channel entrance--i.e., an inner site on-axis and an outer site off-axis. We conclude that the MD-PMF approach is a powerful tool for understanding and predicting the function of narrow ion channels in a manner that is consistent with the atomic and thermally fluctuating nature of proteins.
Figures






Similar articles
-
Molecular dynamics - potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels.Biophys Chem. 2006 Dec 1;124(3):251-67. doi: 10.1016/j.bpc.2006.04.015. Epub 2006 May 9. Biophys Chem. 2006. PMID: 16781050
-
The role of the dielectric barrier in narrow biological channels: a novel composite approach to modeling single-channel currents.Biophys J. 2003 Jun;84(6):3646-61. doi: 10.1016/S0006-3495(03)75095-4. Biophys J. 2003. PMID: 12770873 Free PMC article.
-
Gramicidin A channel as a test ground for molecular dynamics force fields.Biophys J. 2003 Apr;84(4):2159-68. doi: 10.1016/S0006-3495(03)75022-X. Biophys J. 2003. PMID: 12668425 Free PMC article.
-
Theoretical and computational models of biological ion channels.Q Rev Biophys. 2004 Feb;37(1):15-103. doi: 10.1017/s0033583504003968. Q Rev Biophys. 2004. PMID: 17390604 Review.
-
Gramicidin channels.IEEE Trans Nanobioscience. 2005 Mar;4(1):10-20. doi: 10.1109/tnb.2004.842470. IEEE Trans Nanobioscience. 2005. PMID: 15816168 Review.
Cited by
-
Proton transport behavior through the influenza A M2 channel: insights from molecular simulation.Biophys J. 2007 Nov 15;93(10):3470-9. doi: 10.1529/biophysj.107.105742. Epub 2007 Aug 10. Biophys J. 2007. PMID: 17693473 Free PMC article.
-
Challenges and advances in atomistic simulations of potassium and sodium ion channel gating and permeation.J Physiol. 2019 Feb;597(3):679-698. doi: 10.1113/JP277088. Epub 2018 Dec 19. J Physiol. 2019. PMID: 30471114 Free PMC article. Review.
-
A computational kinetic model of diffusion for molecular systems.J Chem Phys. 2013 Sep 28;139(12):121929. doi: 10.1063/1.4820876. J Chem Phys. 2013. PMID: 24089741 Free PMC article.
-
A constricted opening in Kir channels does not impede potassium conduction.Nat Commun. 2020 Jun 15;11(1):3024. doi: 10.1038/s41467-020-16842-0. Nat Commun. 2020. PMID: 32541684 Free PMC article.
-
The voltage-sensing domain of a hERG1 mutant is a cation-selective channel.Biophys J. 2022 Dec 6;121(23):4585-4599. doi: 10.1016/j.bpj.2022.10.032. Epub 2022 Oct 29. Biophys J. 2022. PMID: 36815709 Free PMC article.
References
-
- Karplus, M. 2002. Molecular dynamics simulations of biomolecules. Acc. Chem. Res. 35:321–323. - PubMed
-
- Roux, B., T. Allen, S. Bernèche, and W. Im. 2004. Theoretical and computational models of biological ion channels. Q. Rev. Biophys. 37:15–103. - PubMed
-
- Lindorff-Larsen, K., R. Best, M. Depristo, C. Dobson, and M. Vendruscolo. 2005. Simultaneous determination of protein structure and dynamics. Nature. 433:128–132. - PubMed
-
- Karplus, M., and J. McCammon. 1981. The internal dynamics of globular proteins. CRC Crit. Rev. Biochem. 9:293–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

