HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status
- PMID: 16500994
- PMCID: PMC1435809
- DOI: 10.1104/pp.105.073486
HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status
Abstract
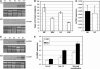
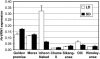
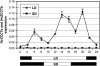
Two genetic loci control the vernalization response in winter cereals; VRN1, which encodes an AP1-like MADS-box transcription factor, and VRN2, which has been mapped to a chromosome region containing ZCCT zinc finger transcription factor genes. We examined whether daylength regulates expression of HvVRN1 and HvVRN2. In a vernalization-responsive winter barley (Hordeum vulgare), expression of HvVRN1 is regulated by vernalization and by development, but not by daylength. Daylength affected HvVRN1 expression in only one of six vernalization-insensitive spring barleys examined and so cannot be a general feature of regulation of this gene. In contrast, daylength is the major determinant of expression levels of two ZCCT genes found at the barley VRN2 locus, HvZCCTa and HvZCCTb. In winter barley, high levels of HvZCCTa and HvZCCTb expression were detected only when plants were grown in long days. During vernalization in long-day conditions, HvVRN1 is induced and expression of HvZCCTb is repressed. During vernalization under short days, induction of HvVRN1 occurs without changes in HvZCCTa and HvZCCTb expression. Analysis of HvZCCTa and HvZCCTb expression levels in a doubled haploid population segregating for different vernalization and daylength requirements showed that HvVRN1 genotype determines HvZCCTa and HvZCCTb expression levels. We conclude that the vernalization response is mediated through HvVRN1, whereas HvZCCTa and HvZCCTb respond to daylength cues to repress flowering under long days in nonvernalized plants.
Figures





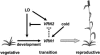
References
-
- Ausubel FM, Brenton R, Kingston RE, Moore DD, Siedman JG, Smyth JA, Struhl K (1994) Current Protocols in Molecular Biology. John Wiley and Sons, New York
-
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 - PubMed
-
- Chang S, Puryear J, Cairney K (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

