Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model
- PMID: 16501048
- PMCID: PMC1450150
- DOI: 10.1073/pnas.0508917103
Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model
Abstract
Duchenne muscular dystrophy is an X-linked muscle disease characterized by mutations in the dystrophin gene. Many of these can be corrected at the posttranscriptional level by skipping the mutated exon. We have obtained persistent exon skipping in mdx mice by tail vein injection with an adeno-associated viral (AAV) vector expressing antisense sequences as part of the stable cellular U1 small nuclear RNA. Systemic delivery of the AAV construct resulted in effective body-wide colonization, significant recovery of the functional properties in vivo, and lower creatine kinase serum levels, suggesting an overall decrease in muscle wasting. The transduced muscles rescued dystrophin expression and displayed a significant recovery of function toward the normal values at single muscle fiber level. This approach provides solid bases for a systemic use of AAV-mediated antisense-U1 small nuclear RNA expression for the therapeutic treatment of Duchenne muscular dystrophy.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
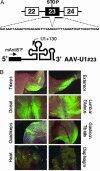
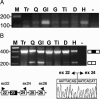

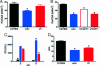
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

