Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function
- PMID: 16501070
- PMCID: PMC1395466
- DOI: 10.1128/JVI.80.6.2596-2608.2006
Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function
Abstract
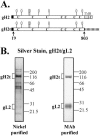
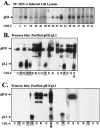
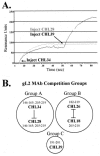
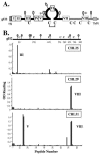
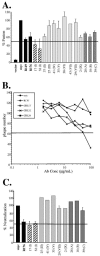
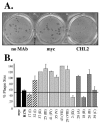
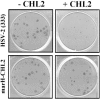
The gH/gL complex plays an essential role in virus entry and cell-cell spread of herpes simplex virus (HSV). Very few immunologic reagents were previously available to either identify important functional regions or gain information about structural features of this complex. Therefore, we generated and characterized a panel of 31 monoclonal antibodies (MAbs) against HSV type 2 (HSV-2) gH/gL. Fourteen MAbs bound to a conformation-dependent epitope of the gH2/gL2 complex, and all blocked virus spread. The other 17 MAbs recognized linear epitopes of gH (12) or gL (5). Interestingly, two of the gL MAbs and six of the gH MAbs were type common. Overlapping synthetic peptides were used to map MAbs against linear epitopes. These data, along with results of competition analyses and functional assays, assigned the MAbs to groups representing eight distinct antigenic sites on gH (I to VIII) and three sites on gL (A, B, and C). Of most importance, the MAbs with biological activity mapped either to site I of gH2 (amino acids 19 to 38) or to sites B and C of gL2 (residues 191 to 210). Thus, these MAbs constitute a novel set of reagents, including the first such reagents against gH2 and gL2 as well as some that recognize both serotypes of each protein. Several recognize important functional domains of gH2, gL2, or the complex. We suggest a common grouping scheme for all of the known MAbs against gH/gL of both HSV-1 and HSV-2.
Figures


Similar articles
-
Using Antibodies and Mutants To Localize the Presumptive gH/gL Binding Site on Herpes Simplex Virus gD.J Virol. 2018 Nov 27;92(24):e01694-18. doi: 10.1128/JVI.01694-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30282715 Free PMC article.
-
Localization of type-specific epitopes of herpes simplex virus type 2 glycoprotein G recognized by human and mouse antibodies.J Gen Virol. 1998 May;79 ( Pt 5):1215-24. doi: 10.1099/0022-1317-79-5-1215. J Gen Virol. 1998. PMID: 9603337
-
Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex.J Virol. 1998 Jul;72(7):6092-103. doi: 10.1128/JVI.72.7.6092-6103.1998. J Virol. 1998. PMID: 9621073 Free PMC article.
-
Antigenic and structural properties of mutants in herpes simplex virus 1 glycoprotein B.Adv Exp Med Biol. 1990;278:165-82. doi: 10.1007/978-1-4684-5853-4_17. Adv Exp Med Biol. 1990. PMID: 1705078 Review. No abstract available.
-
Stuck in the middle: structural insights into the role of the gH/gL heterodimer in herpesvirus entry.Curr Opin Virol. 2013 Feb;3(1):13-9. doi: 10.1016/j.coviro.2012.10.005. Epub 2012 Oct 26. Curr Opin Virol. 2013. PMID: 23107819 Free PMC article. Review.
Cited by
-
A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL.Proc Natl Acad Sci U S A. 2015 May 12;112(19):6056-61. doi: 10.1073/pnas.1501176112. Epub 2015 Apr 27. Proc Natl Acad Sci U S A. 2015. PMID: 25918416 Free PMC article.
-
Regulation of Herpes Simplex Virus Glycoprotein-Induced Cascade of Events Governing Cell-Cell Fusion.J Virol. 2016 Nov 14;90(23):10535-10544. doi: 10.1128/JVI.01501-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27630245 Free PMC article.
-
Mutations in the amino terminus of herpes simplex virus type 1 gL can reduce cell-cell fusion without affecting gH/gL trafficking.J Virol. 2014 Jan;88(1):739-44. doi: 10.1128/JVI.02383-13. Epub 2013 Oct 23. J Virol. 2014. PMID: 24155377 Free PMC article.
-
Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors.mBio. 2013 Feb 26;4(2):e00046-13. doi: 10.1128/mBio.00046-13. mBio. 2013. PMID: 23443004 Free PMC article.
-
Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B.J Virol. 2007 May;81(9):4858-65. doi: 10.1128/JVI.02755-06. Epub 2007 Feb 21. J Virol. 2007. PMID: 17314168 Free PMC article.
References
-
- Anderson, R. A., D. X. Liu, and U. A. Gompels. 1996. Definition of a human herpesvirus-6 betaherpesvirus-specific domain in glycoprotein gH that governs interaction with glycoprotein gL: substitution of human cytomegalovirus glycoproteins permits group-specific complex formation. Virology 217:517-526. - PubMed
-
- Buckmaster, E. A., U. Gompels, and A. Minson. 1984. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 10(3) molecular weight. Virology 139:408-413. - PubMed
-
- Cairns, T. M., D. J. Landsburg, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332:550-562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

