Identification and characterization of an exogenous retrovirus from atlantic salmon swim bladder sarcomas
- PMID: 16501103
- PMCID: PMC1395439
- DOI: 10.1128/JVI.80.6.2941-2948.2006
Identification and characterization of an exogenous retrovirus from atlantic salmon swim bladder sarcomas
Abstract
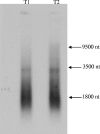
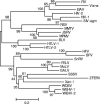
A novel piscine retrovirus has been identified in association with an outbreak of leiomyosarcoma in the swim bladders of Atlantic salmon. The complete nucleotide sequence of the Atlantic salmon swim bladder sarcoma virus (SSSV) provirus is 10.9 kb in length and shares a structure and transcriptional profile similar to those of murine leukemia virus-like simple retroviruses. SSSV appears unique to simple retroviruses by not harboring sequences in the Atlantic salmon genome. Additionally, SSSV differs from other retroviruses in potentially utilizing a methionine tRNA primer binding site. SSSV-associated tumors contain high proviral copy numbers (greater than 30 per cell) and a polyclonal integration pattern. Phylogenetic analysis based on reverse transcriptase places SSSV with zebrafish endogenous retrovirus (ZFERV) between the Gammaretrovirus and Epsilonretrovirus genera. Large regions of continuous homology between SSSV and ZFERV Gag, Pol, and Env suggest that these viruses represent a new group of related piscine retroviruses.
Figures



Similar articles
-
Swimbladder Leiomyosarcoma in Atlantic salmon (Salmo salar) in North America.J Wildl Dis. 2012 Jul;48(3):795-8. doi: 10.7589/0090-3558-48.3.795. J Wildl Dis. 2012. PMID: 22740549
-
Pathology of tumors in fish associated with retroviruses: a review.Vet Pathol. 2013 May;50(3):390-403. doi: 10.1177/0300985813480529. Epub 2013 Mar 1. Vet Pathol. 2013. PMID: 23456970 Review.
-
Sequence and transcriptional analyses of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: evidence for a gene duplication.J Virol. 1999 Nov;73(11):9393-403. doi: 10.1128/JVI.73.11.9393-9403.1999. J Virol. 1999. PMID: 10516048 Free PMC article.
-
Complete nucleotide sequence and transcriptional analysis of snakehead fish retrovirus.J Virol. 1996 Jun;70(6):3606-16. doi: 10.1128/JVI.70.6.3606-3616.1996. J Virol. 1996. PMID: 8648695 Free PMC article.
-
Molecular biology of type A endogenous retrovirus.Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review.
Cited by
-
Peptides derived from HIV-1 integrase that bind Rev stimulate viral genome integration.PLoS One. 2009;4(1):e4155. doi: 10.1371/journal.pone.0004155. Epub 2009 Jan 7. PLoS One. 2009. PMID: 19127291 Free PMC article.
-
Infection and cancer in multicellular organisms.Philos Trans R Soc Lond B Biol Sci. 2015 Jul 19;370(1673):20140224. doi: 10.1098/rstb.2014.0224. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26056368 Free PMC article. Review.
-
Walleye dermal sarcoma virus: molecular biology and oncogenesis.Viruses. 2010 Sep;2(9):1984-1999. doi: 10.3390/v2091984. Epub 2010 Sep 22. Viruses. 2010. PMID: 21994717 Free PMC article.
-
Multiple groups of endogenous epsilon-like retroviruses conserved across primates.J Virol. 2014 Nov;88(21):12464-71. doi: 10.1128/JVI.00966-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142585 Free PMC article.
-
Characterizing novel endogenous retroviruses from genetic variation inferred from short sequence reads.Sci Rep. 2015 Oct 23;5:15644. doi: 10.1038/srep15644. Sci Rep. 2015. PMID: 26493184 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Blum, H. E., J. D. Harris, P. Ventura, D. Walker, K. Staskus, E. Retzel, and A. T. Haase. 1985. Synthesis in cell culture of the gapped linear duplex DNA of the slow virus visna. Virology 142:270-277. - PubMed
-
- Bowser, P. R., J. W. Casey, R. N. Casey, S. L. Quackenbush, L. Lofton, J. A. Coll, and R. C. Cipriano. Submitted for publication.
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

