A system for automated lexical mapping
- PMID: 16501186
- PMCID: PMC1513662
- DOI: 10.1197/jamia.M1823
A system for automated lexical mapping
Abstract
Objective: To automate the mapping of disparate databases to standardized medical vocabularies.
Background: Merging of clinical systems and medical databases, or aggregation of information from disparate databases, frequently requires a process whereby vocabularies are compared and similar concepts are mapped.
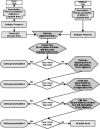
Design: Using a normalization phase followed by a novel alignment stage inspired by DNA sequence alignment methods, automated lexical mapping can map terms from various databases to standard vocabularies such as the UMLS (Unified Medical Language System) and LOINC (Logical Observation Identifier Names and Codes).
Measurements: This automated lexical mapping was evaluated using three real-world laboratory databases from different health care institutions. The authors report the sensitivity, specificity, percentage correct (true positives plus true negatives divided by total number of terms), and true positive and true negative rates as measures of system performance.
Results: The alignment algorithm was able to map 57% to 78% (average of 63% over all runs and databases) of equivalent concepts through lexical mapping alone. True positive rates ranged from 18% to 70%; true negative rates ranged from 5% to 52%.
Conclusion: Lexical mapping can facilitate the integration of data from diverse sources and decrease the time and cost required for manual mapping and integration of clinical systems and medical databases.
Figures


Similar articles
-
Methods for automated concept mapping between medical databases.J Biomed Inform. 2004 Jun;37(3):162-78. doi: 10.1016/j.jbi.2004.03.003. J Biomed Inform. 2004. PMID: 15196481
-
An automated approach to mapping external terminologies to the UMLS.IEEE Trans Biomed Eng. 2009 Jun;56(6):1598-605. doi: 10.1109/TBME.2009.2015651. Epub 2009 Mar 4. IEEE Trans Biomed Eng. 2009. PMID: 19272981
-
Automating identification of adverse events related to abnormal lab results using standard vocabularies.AMIA Annu Symp Proc. 2005;2005:903. AMIA Annu Symp Proc. 2005. PMID: 16779190 Free PMC article.
-
Logical Observation Identifiers Names and Codes for Laboratorians.Arch Pathol Lab Med. 2020 Feb;144(2):229-239. doi: 10.5858/arpa.2018-0477-RA. Epub 2019 Jun 20. Arch Pathol Lab Med. 2020. PMID: 31219342 Review.
-
Choosing Among Common Data Models for Real-World Data Analyses Fit for Making Decisions About the Effectiveness of Medical Products.Clin Pharmacol Ther. 2020 Apr;107(4):827-833. doi: 10.1002/cpt.1577. Epub 2019 Aug 25. Clin Pharmacol Ther. 2020. PMID: 31330042 Review.
Cited by
-
A corpus-based approach for automated LOINC mapping.J Am Med Inform Assoc. 2014 Jan-Feb;21(1):64-72. doi: 10.1136/amiajnl-2012-001159. Epub 2013 May 15. J Am Med Inform Assoc. 2014. PMID: 23676247 Free PMC article.
-
Translating the Foundational Model of Anatomy into French using knowledge-based and lexical methods.BMC Med Inform Decis Mak. 2011 Oct 26;11:65. doi: 10.1186/1472-6947-11-65. BMC Med Inform Decis Mak. 2011. PMID: 22029629 Free PMC article.
-
A computational linguistics motivated mapping of ICPC-2 PLUS to SNOMED CT.BMC Med Inform Decis Mak. 2008 Oct 27;8 Suppl 1(Suppl 1):S5. doi: 10.1186/1472-6947-8-S1-S5. BMC Med Inform Decis Mak. 2008. PMID: 19007442 Free PMC article.
-
Free and open source enabling technologies for patient-centric, guideline-based clinical decision support: a survey.Yearb Med Inform. 2007:74-86. Yearb Med Inform. 2007. PMID: 17700908 Free PMC article.
-
Learning From the Crowd in Terminology Mapping: The LOINC Experience.Lab Med. 2015 Spring;46(2):168-74. doi: 10.1309/LMWJ730SVKTUBAOJ. Lab Med. 2015. PMID: 25918199 Free PMC article.
References
-
- Elkin PL, Cimino JJ, Lowe HJ, Aronow DB, Payne TH, Pincetl PS, Barnett GO. Mapping to MeSH: the art of trapping MeSH equivalence from within narrative text. Proc 12th SCAMC. 1988:185–90.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

