The hybrid state of tRNA binding is an authentic translation elongation intermediate
- PMID: 16501572
- PMCID: PMC1687179
- DOI: 10.1038/nsmb1060
The hybrid state of tRNA binding is an authentic translation elongation intermediate
Abstract
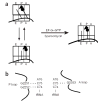
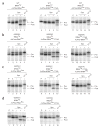
The GTPase elongation factor (EF)-G is responsible for promoting the translocation of the messenger RNA-transfer RNA complex on the ribosome, thus opening up the A site for the next aminoacyl-tRNA. Chemical modification and cryo-EM studies have indicated that tRNAs can bind the ribosome in an alternative 'hybrid' state after peptidyl transfer and before translocation, though the relevance of this state during translation elongation has been a subject of debate. Here, using pre-steady-state kinetic approaches and mutant analysis, we show that translocation by EF-G is most efficient when tRNAs are bound in a hybrid state, supporting the argument that this state is an authentic intermediate during translation.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Figures

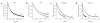
References
-
- Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. - PubMed
-
- Piepenburg O, et al. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. - PubMed
-
- Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

