SNP-RFLPing: restriction enzyme mining for SNPs in genomes
- PMID: 16503968
- PMCID: PMC1386656
- DOI: 10.1186/1471-2164-7-30
SNP-RFLPing: restriction enzyme mining for SNPs in genomes
Abstract
Background: The restriction fragment length polymorphism (RFLP) is a common laboratory method for the genotyping of single nucleotide polymorphisms (SNPs). Here, we describe a web-based software, named SNP-RFLPing, which provides the restriction enzyme for RFLP assays on a batch of SNPs and genes from the human, rat, and mouse genomes.
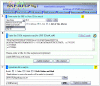
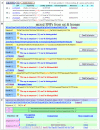
Results: Three user-friendly inputs are included: 1) NCBI dbSNP "rs" or "ss" IDs; 2) NCBI Entrez gene ID and HUGO gene name; 3) any formats of SNP-in-sequence, are allowed to perform the SNP-RFLPing assay. These inputs are auto-programmed to SNP-containing sequences and their complementary sequences for the selection of restriction enzymes. All SNPs with available RFLP restriction enzymes of each input genes are provided even if many SNPs exist. The SNP-RFLPing analysis provides the SNP contig position, heterozygosity, function, protein residue, and amino acid position for cSNPs, as well as commercial and non-commercial restriction enzymes.
Conclusion: This web-based software solves the input format problems in similar softwares and greatly simplifies the procedure for providing the RFLP enzyme. Mixed free forms of input data are friendly to users who perform the SNP-RFLPing assay. SNP-RFLPing offers a time-saving application for association studies in personalized medicine and is freely available at http://bio.kuas.edu.tw/snp-rflp/.
Figures

References
-
- Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. http://tools.neb.com/NEBcutter2/index.php - DOI - PMC - PubMed
-
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005:D230–232. http://rebase.neb.com/rebase/rebase.html - PMC - PubMed
-
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. http://www.ncbi.nlm.nih.gov/projects/SNP/ - DOI - PMC - PubMed
-
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005:D54–58. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene - PMC - PubMed
-
- Charras C, Lecroq T. Handbook of Exact String Matching Algorithms. King's College Publications; 2004. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

