The PowerAtlas: a power and sample size atlas for microarray experimental design and research
- PMID: 16504070
- PMCID: PMC1395338
- DOI: 10.1186/1471-2105-7-84
The PowerAtlas: a power and sample size atlas for microarray experimental design and research
Abstract
Background: Microarrays permit biologists to simultaneously measure the mRNA abundance of thousands of genes. An important issue facing investigators planning microarray experiments is how to estimate the sample size required for good statistical power. What is the projected sample size or number of replicate chips needed to address the multiple hypotheses with acceptable accuracy? Statistical methods exist for calculating power based upon a single hypothesis, using estimates of the variability in data from pilot studies. There is, however, a need for methods to estimate power and/or required sample sizes in situations where multiple hypotheses are being tested, such as in microarray experiments. In addition, investigators frequently do not have pilot data to estimate the sample sizes required for microarray studies.
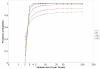
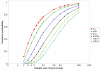
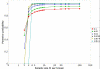
Results: To address this challenge, we have developed a Microrarray PowerAtlas. The atlas enables estimation of statistical power by allowing investigators to appropriately plan studies by building upon previous studies that have similar experimental characteristics. Currently, there are sample sizes and power estimates based on 632 experiments from Gene Expression Omnibus (GEO). The PowerAtlas also permits investigators to upload their own pilot data and derive power and sample size estimates from these data. This resource will be updated regularly with new datasets from GEO and other databases such as The Nottingham Arabidopsis Stock Center (NASC).
Conclusion: This resource provides a valuable tool for investigators who are planning efficient microarray studies and estimating required sample sizes.
Figures









References
-
- PowerAtlas T. http://www.poweratlas.org. 2006. http://www.poweratlas.org
-
- Gadbury GL, Page GP, Edwards J, Kayo T, Weindruch R, Permana PA, Mountz J, Allison DB. Power Analysis and Sample Size Estimation in the Age of High Dimensional Biology. Stat Meth Med Res. 2004;13:325–338.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

