Bias in error estimation when using cross-validation for model selection
- PMID: 16504092
- PMCID: PMC1397873
- DOI: 10.1186/1471-2105-7-91
Bias in error estimation when using cross-validation for model selection
Abstract
Background: Cross-validation (CV) is an effective method for estimating the prediction error of a classifier. Some recent articles have proposed methods for optimizing classifiers by choosing classifier parameter values that minimize the CV error estimate. We have evaluated the validity of using the CV error estimate of the optimized classifier as an estimate of the true error expected on independent data.
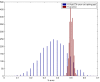
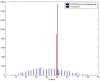
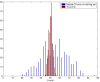
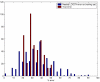
Results: We used CV to optimize the classification parameters for two kinds of classifiers; Shrunken Centroids and Support Vector Machines (SVM). Random training datasets were created, with no difference in the distribution of the features between the two classes. Using these "null" datasets, we selected classifier parameter values that minimized the CV error estimate. 10-fold CV was used for Shrunken Centroids while Leave-One-Out-CV (LOOCV) was used for the SVM. Independent test data was created to estimate the true error. With "null" and "non null" (with differential expression between the classes) data, we also tested a nested CV procedure, where an inner CV loop is used to perform the tuning of the parameters while an outer CV is used to compute an estimate of the error. The CV error estimate for the classifier with the optimal parameters was found to be a substantially biased estimate of the true error that the classifier would incur on independent data. Even though there is no real difference between the two classes for the "null" datasets, the CV error estimate for the Shrunken Centroid with the optimal parameters was less than 30% on 18.5% of simulated training data-sets. For SVM with optimal parameters the estimated error rate was less than 30% on 38% of "null" data-sets. Performance of the optimized classifiers on the independent test set was no better than chance. The nested CV procedure reduces the bias considerably and gives an estimate of the error that is very close to that obtained on the independent testing set for both Shrunken Centroids and SVM classifiers for "null" and "non-null" data distributions.
Conclusion: We show that using CV to compute an error estimate for a classifier that has itself been tuned using CV gives a significantly biased estimate of the true error. Proper use of CV for estimating true error of a classifier developed using a well defined algorithm requires that all steps of the algorithm, including classifier parameter tuning, be repeated in each CV loop. A nested CV procedure provides an almost unbiased estimate of the true error.
Figures
Similar articles
-
Classification based upon gene expression data: bias and precision of error rates.Bioinformatics. 2007 Jun 1;23(11):1363-70. doi: 10.1093/bioinformatics/btm117. Epub 2007 Mar 28. Bioinformatics. 2007. PMID: 17392326 Review.
-
Prediction error estimation: a comparison of resampling methods.Bioinformatics. 2005 Aug 1;21(15):3301-7. doi: 10.1093/bioinformatics/bti499. Epub 2005 May 19. Bioinformatics. 2005. PMID: 15905277
-
What should be expected from feature selection in small-sample settings.Bioinformatics. 2006 Oct 1;22(19):2430-6. doi: 10.1093/bioinformatics/btl407. Epub 2006 Jul 26. Bioinformatics. 2006. PMID: 16870934
-
Improved centroids estimation for the nearest shrunken centroid classifier.Bioinformatics. 2007 Apr 15;23(8):972-9. doi: 10.1093/bioinformatics/btm046. Epub 2007 Mar 24. Bioinformatics. 2007. PMID: 17384429
-
Support vector machine applications in bioinformatics.Appl Bioinformatics. 2003;2(2):67-77. Appl Bioinformatics. 2003. PMID: 15130823 Review.
Cited by
-
Activating and relaxing music entrains the speed of beat synchronized walking.PLoS One. 2013 Jul 10;8(7):e67932. doi: 10.1371/journal.pone.0067932. Print 2013. PLoS One. 2013. PMID: 23874469 Free PMC article. Clinical Trial.
-
Utilization of machine learning to test the impact of cognitive processing and emotion recognition on the development of PTSD following trauma exposure.BMC Psychiatry. 2020 Jun 23;20(1):325. doi: 10.1186/s12888-020-02728-4. BMC Psychiatry. 2020. PMID: 32576245 Free PMC article.
-
Using machine learning with intensive longitudinal data to predict depression and suicidal ideation among medical interns over time.Psychol Med. 2023 Sep;53(12):5778-5785. doi: 10.1017/S0033291722003014. Epub 2022 Sep 30. Psychol Med. 2023. PMID: 36177889 Free PMC article.
-
Cross-validation of predictive models for functional recovery after post-stroke rehabilitation.J Neuroeng Rehabil. 2022 Sep 7;19(1):96. doi: 10.1186/s12984-022-01075-7. J Neuroeng Rehabil. 2022. PMID: 36071452 Free PMC article.
-
Genomic biomarkers for personalized medicine: development and validation in clinical studies.Comput Math Methods Med. 2013;2013:865980. doi: 10.1155/2013/865980. Epub 2013 Apr 17. Comput Math Methods Med. 2013. PMID: 23690882 Free PMC article. Review.
References
-
- Duda RO, Hart PE, Stork DG. Pattern classification. John Wiley and Sons Inc. 2001;Ch.9:483–486.
-
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. - PubMed
-
- Reunanen J. Overfitting in making comparisons between variable selection methods. J Machine Learning Research. 2003;3:1371–1382. doi: 10.1162/153244303322753715. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

