A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition
- PMID: 16505140
- PMCID: PMC2118261
- DOI: 10.1084/jem.20051777
A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition
Abstract
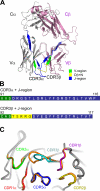
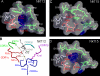
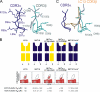
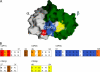
Little is known regarding the basis for selection of the semi-invariant alphabeta T cell receptor (TCR) expressed by natural killer T (NKT) cells or how this mediates recognition of CD1d-glycolipid complexes. We have determined the structures of two human NKT TCRs that differ in their CDR3beta composition and length. Both TCRs contain a conserved, positively charged pocket at the ligand interface that is lined by residues from the invariant TCR alpha- and semi-invariant beta-chains. The cavity is centrally located and ideally suited to interact with the exposed glycosyl head group of glycolipid antigens. Sequences common to mouse and human invariant NKT TCRs reveal a contiguous conserved "hot spot" that provides a basis for the reactivity of NKT cells across species. Structural and functional data suggest that the CDR3beta loop provides a plasticity mechanism that accommodates recognition of a variety of glycolipid antigens presented by CD1d. We propose a model of NKT TCR-CD1d-glycolipid interaction in which the invariant CDR3alpha loop is predicted to play a major role in determining the inherent bias toward CD1d. The findings define a structural basis for the selection of the semi-invariant alphabeta TCR and the unique antigen specificity of NKT cells.
Figures

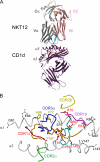
References
-
- Ulrichs, T., and S.A. Porcelli. 2000. CD1 proteins: targets of T cell recognition in innate and adaptive immunity. Rev. Immunogenet. 2:416–432. - PubMed
-
- Brigl, M., and M.B. Brenner. 2004. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 22:817–890. - PubMed
-
- Kobayashi, E., K. Motoki, T. Uchida, H. Fukushima, and Y. Koezuka. 1995. Krn7000, a novel immunomodulator, and its antitumor activities. Oncol. Res. 7:529–534. - PubMed
-
- Hansen, D.S., M.A. Siomos, L. Buckingham, A.A. Scalzo, and L. Schofield. 2003. Regulation of murine cerebral malaria pathogenesis by CD1d-restricted NKT cells and the natural killer complex. Immunity. 18:391–402. - PubMed
-
- Schofield, L., M.J. McConville, D. Hansen, A.S. Campbell, B. Fraser-Reid, M.J. Grusby, and S.D. Tachado. 1999. CD1d-restricted immunoglobulin G formation to GPI-anchored antigens mediated by NKT cells. Science. 283:225–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

