Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1
- PMID: 16505166
- PMCID: PMC2063703
- DOI: 10.1083/jcb.200508166
Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1
Abstract
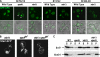
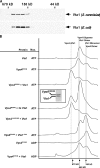
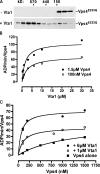
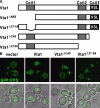
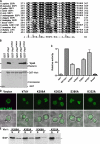
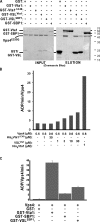
In eukaryotes, the multivesicular body (MVB) sorting pathway plays an essential role in regulating cell surface protein composition, thereby impacting numerous cellular functions. Vps4, an ATPase associated with a variety of cellular activities, is required late in the MVB sorting reaction to dissociate the endosomal sorting complex required for transport (ESCRT), a requisite for proper function of this pathway. However, regulation of Vps4 function is not understood. We characterize Vta1 as a positive regulator of Vps4 both in vivo and in vitro. Vta1 promotes proper assembly of Vps4 and stimulates its ATPase activity through the conserved Vta1/SBP1/LIP5 region present in Vta1 homologues across evolution, including human SBP1 and Arabidopsis thaliana LIP5. These results suggest an evolutionarily conserved mechanism through which the disassembly of the ESCRT proteins, and thereby MVB sorting, is regulated by the Vta1/SBP1/LIP5 proteins.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

