Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide
- PMID: 16505360
- PMCID: PMC1450129
- DOI: 10.1073/pnas.0600082103
Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide
Abstract
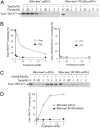
Studies in vitro have established that free tryptophan induces tna operon expression by binding to the ribosome that has just completed synthesis of TnaC-tRNA(Pro), the peptidyl-tRNA precursor of the leader peptide of this operon. Tryptophan acts by inhibiting Release Factor 2-mediated cleavage of this peptidyl-tRNA at the tnaC stop codon. Here we analyze the ribosomal location of free tryptophan, the changes it produces in the ribosome, and the role of the nascent TnaC-tRNA(Pro) peptide in facilitating tryptophan binding and induction. The positional changes of 23S rRNA nucleotides that occur during induction were detected by using methylation protection and binding/competition assays. The ribosome-TnaC-tRNA(Pro) complexes analyzed were formed in vitro; they contained either wild-type TnaC-tRNA(Pro) or its nonfunctional substitute, TnaC(W12R)-tRNA(Pro). Upon comparing these two peptidyl-tRNA-ribosome complexes, free tryptophan was found to block methylation of nucleotide A2572 of wild-type ribosome-TnaC-tRNA(Pro) complexes but not of ribosome-TnaC(W12R)-tRNA(Pro) complexes. Nucleotide A2572 is in the ribosomal peptidyl transferase center. Tryptophanol, a noninducing competitor of tryptophan, was ineffective in blocking A2572 methylation; however, it did reverse the protective effect of tryptophan. Free tryptophan inhibited puromycin cleavage of TnaC-tRNA(Pro); it also inhibited binding of the antibiotic sparsomycin. These effects were not observed with TnaC(W12R)-tRNA(Pro) mutant complexes. These findings establish that Trp-12 of TnaC-tRNA(Pro) is required for introducing specific changes in the peptidyl transferase center of the ribosome that activate free tryptophan binding, resulting in peptidyl transferase inhibition. Free tryptophan appears to act at or near the binding sites of several antibiotics in the peptidyl transferase center.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures




References
-
- Gong F., Yanofsky C. J. Biol. Chem. 2002;277:17095–17100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

