The colorectal microRNAome
- PMID: 16505370
- PMCID: PMC1450142
- DOI: 10.1073/pnas.0511155103
The colorectal microRNAome
Abstract
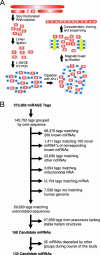
MicroRNAs (miRNAs) are a class of small noncoding RNAs that have important regulatory roles in multicellular organisms. The public miRNA database contains 321 human miRNA sequences, 234 of which have been experimentally verified. To explore the possibility that additional miRNAs are present in the human genome, we have developed an experimental approach called miRNA serial analysis of gene expression (miRAGE) and used it to perform the largest experimental analysis of human miRNAs to date. Sequence analysis of 273,966 small RNA tags from human colorectal cells allowed us to identify 200 known mature miRNAs, 133 novel miRNA candidates, and 112 previously uncharacterized miRNA* forms. To aid in the evaluation of candidate miRNAs, we disrupted the Dicer locus in three human colorectal cancer cell lines and examined known and novel miRNAs in these cells. These studies suggest that the human genome contains many more miRNAs than currently identified and provide an approach for the large-scale experimental cloning of novel human miRNAs in human tissues.
Conflict of interest statement
Conflict of interest statement: K.W.K. and V.E.V. receive research funding from Genzyme, and K.W.K., V.E.V., and Johns Hopkins University own Genzyme stock, which is subject to certain restrictions under Johns Hopkins University policy. K.W.K. and V.E.V. are also paid consultants to Genzyme. The terms of this arrangement are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Figures




Similar articles
-
Identification of microRNA in the protist Trichomonas vaginalis.Genomics. 2009 May;93(5):487-93. doi: 10.1016/j.ygeno.2009.01.004. Epub 2009 Feb 3. Genomics. 2009. PMID: 19442639
-
Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1881-9. doi: 10.1073/pnas.1602532113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976605 Free PMC article.
-
The regulatory epicenter of miRNAs.J Biosci. 2011 Sep;36(4):621-38. doi: 10.1007/s12038-011-9109-y. J Biosci. 2011. PMID: 21857109
-
Role of miRNA in carcinogenesis and biomarker selection: a methodological view.Expert Rev Mol Diagn. 2007 Sep;7(5):569-603. doi: 10.1586/14737159.7.5.569. Expert Rev Mol Diagn. 2007. PMID: 17892365 Review.
-
MicroRNAs in oligodendrocyte and Schwann cell differentiation.Dev Neurosci. 2011;33(1):14-20. doi: 10.1159/000323919. Epub 2011 Feb 23. Dev Neurosci. 2011. PMID: 21346322 Free PMC article. Review.
Cited by
-
MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice.Gastroenterology. 2013 Aug;145(2):437-46. doi: 10.1053/j.gastro.2013.04.012. Epub 2013 Apr 22. Gastroenterology. 2013. PMID: 23619147 Free PMC article.
-
The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes.Biology (Basel). 2023 Dec 18;12(12):1536. doi: 10.3390/biology12121536. Biology (Basel). 2023. PMID: 38132362 Free PMC article.
-
MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity.Oncogene. 2016 Jan 28;35(4):448-58. doi: 10.1038/onc.2015.96. Epub 2015 Apr 13. Oncogene. 2016. PMID: 25867061 Free PMC article. Clinical Trial.
-
Radiation-induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb.Oncotarget. 2015 Jun 30;6(18):15984-94. doi: 10.18632/oncotarget.3762. Oncotarget. 2015. PMID: 25961730 Free PMC article.
-
MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer.Neoplasia. 2012 Sep;14(9):868-79. doi: 10.1593/neo.121094. Neoplasia. 2012. PMID: 23019418 Free PMC article.
References
-
- Bartel D. P. Cell. 2004;116:281–297. - PubMed
-
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. Nature. 2001;409:363–366. - PubMed
-
- Lim L. P., Glasner M. E., Yekta S., Burge C. B., Bartel D. P. Science. 2003;299:1540. - PubMed
-
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Nat. Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

