Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors
- PMID: 16505381
- PMCID: PMC1383502
- DOI: 10.1073/pnas.0508214103
Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors
Abstract
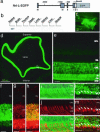
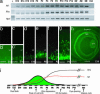
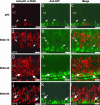
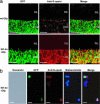
The Maf-family transcription factor Nrl is a key regulator of photoreceptor differentiation in mammals. Ablation of the Nrl gene in mice leads to functional cones at the expense of rods. We show that a 2.5-kb Nrl promoter segment directs the expression of enhanced GFP specifically to rod photoreceptors and the pineal gland of transgenic mice. GFP is detected shortly after terminal cell division, corresponding to the timing of rod genesis revealed by birthdating studies. In Nrl-/- retinas, the GFP+ photoreceptors express S-opsin, consistent with the transformation of rod precursors into cones. We report the gene profiles of freshly isolated flow-sorted GFP+ photoreceptors from wild-type and Nrl-/- retinas at five distinct developmental stages. Our results provide a framework for establishing gene regulatory networks that lead to mature functional photoreceptors from postmitotic precursors. Differentially expressed rod and cone genes are excellent candidates for retinopathies.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

