HIV-1 protease flaps spontaneously close to the correct structure in simulations following manual placement of an inhibitor into the open state
- PMID: 16506755
- PMCID: PMC2555982
- DOI: 10.1021/ja058211x
HIV-1 protease flaps spontaneously close to the correct structure in simulations following manual placement of an inhibitor into the open state
Abstract
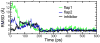
We report unrestrained, all-atom molecular dynamics simulations of HIV-1 protease (HIV-PR) with a continuum solvent model that reproducibly sample closing of the active site flaps following manual placement of a cyclic urea inhibitor into the substrate binding site of the open protease. The open form was obtained from the unbound, semi-open HIV-PR crystal structure, which we recently reported (Hornak, V.; et al. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 915-920.) to have spontaneously opened during unrestrained dynamics. In those simulations, the transiently open flaps always returned to the semi-open form that is observed in all crystal structures of the free protease. Here, we show that manual docking of the inhibitor reproducibly induces spontaneous conversion to the closed form as seen in all inhibitor-bound HIV-PR crystal structures. These simulations reproduced not only the greater degree of flap closure, but also the striking difference in flap "handedness" between bound and free enzyme. In most of the simulations, the final structures were highly accurate. Root-mean-square deviations (RMSD) from the crystal structure of the complex were approximately 1.5 A (averaged over the last 100 ps) for the inhibitor and each flap despite initial RMSD of 2-5 A for the inhibitors and 6-11 A for the flaps. Key hydrogen bonds were formed between the flap tips and between flaps and inhibitor that match those seen in the crystal structure. The results demonstrate that all-atom simulations have the ability to significantly improve poorly docked ligand conformations and reproduce large-scale receptor conformational changes that occur upon binding.
Figures
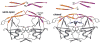

References
-
- Vondrasek J, Wlodawer A. Proteins-Structure Function and Genetics. 2002;49:429–431. - PubMed
-
- Wlodawer A, Vondrasek J. Annual Review of Biophysics and Biomolecular Structure. 1998;27:249–284. - PubMed
-
- Onufriev A, Bashford D, Case DA. Journal of Physical Chemistry B. 2000;104:3712–3720.
-
- Still WC, Tempczyk A, Hawley RC, Hendrickson T. Journal of the American Chemical Society. 1990;112:6127–6129.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

