The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries
- PMID: 16507094
- PMCID: PMC1420335
- DOI: 10.1186/1471-2105-7-97
The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries
Abstract
Background: With the vast amounts of biomedical data being generated by high-throughput analysis methods, controlled vocabularies and ontologies are becoming increasingly important to annotate units of information for ease of search and retrieval. Each scientific community tends to create its own locally available ontology. The interfaces to query these ontologies tend to vary from group to group. We saw the need for a centralized location to perform controlled vocabulary queries that would offer both a lightweight web-accessible user interface as well as a consistent, unified SOAP interface for automated queries.
Results: The Ontology Lookup Service (OLS) was created to integrate publicly available biomedical ontologies into a single database. All modified ontologies are updated daily. A list of currently loaded ontologies is available online. The database can be queried to obtain information on a single term or to browse a complete ontology using AJAX. Auto-completion provides a user-friendly search mechanism. An AJAX-based ontology viewer is available to browse a complete ontology or subsets of it. A programmatic interface is available to query the webservice using SOAP. The service is described by a WSDL descriptor file available online. A sample Java client to connect to the webservice using SOAP is available for download from SourceForge. All OLS source code is publicly available under the open source Apache Licence.
Conclusion: The OLS provides a user-friendly single entry point for publicly available ontologies in the Open Biomedical Ontology (OBO) format. It can be accessed interactively or programmatically at http://www.ebi.ac.uk/ontology-lookup/.
Figures

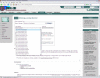

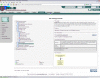
References
-
- Orchard S, Montecchi-Palazzi L, Hermjakob H, Apweiler R. The use of common ontologies and controlled vocabularies to enable data exchange and deposition for complex proteomic experiments. In: Altman, RB, editor. Proceedings of the Pacific Symposium on Biocomputing: 4–8 January 2005; Hawaii. World Press; 2005. pp. 186–96. - PubMed
-
- Open Biomedical Ontologies http://obo.sourceforge.net/
-
- The Gene Ontology http://www.geneontology.org/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

