Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana
- PMID: 16507168
- PMCID: PMC1431724
- DOI: 10.1186/gb-2006-7-2-r13
Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana
Abstract
Background: Genome analyses have revealed that gene duplication in plants is rampant. Furthermore, many of the duplicated genes seem to have been created through ancient genome-wide duplication events. Recently, we have shown that gene loss is strikingly different for large- and small-scale duplication events and highly biased towards the functional class to which a gene belongs. Here, we study the expression divergence of genes that were created during large- and small-scale gene duplication events by means of microarray data and investigate both the influence of the origin (mode of duplication) and the function of the duplicated genes on expression divergence.
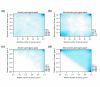
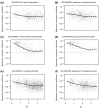
Results: Duplicates that have been created by large-scale duplication events and that can still be found in duplicated segments have expression patterns that are more correlated than those that were created by small-scale duplications or those that no longer lie in duplicated segments. Moreover, the former tend to have highly redundant or overlapping expression patterns and are mostly expressed in the same tissues, while the latter show asymmetric divergence. In addition, a strong bias in divergence of gene expression was observed towards gene function and the biological process genes are involved in.
Conclusion: By using microarray expression data for Arabidopsis thaliana, we show that the mode of duplication, the function of the genes involved, and the time since duplication play important roles in the divergence of gene expression and, therefore, in the functional divergence of genes after duplication.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

