Controlled loading of oligodeoxyribonucleotide monolayers onto unoxidized crystalline silicon; fluorescence-based determination of the surface coverage and of the hybridization efficiency; parallel imaging of the process by Atomic Force Microscopy
- PMID: 16507670
- PMCID: PMC1385995
- DOI: 10.1093/nar/gnj034
Controlled loading of oligodeoxyribonucleotide monolayers onto unoxidized crystalline silicon; fluorescence-based determination of the surface coverage and of the hybridization efficiency; parallel imaging of the process by Atomic Force Microscopy
Abstract
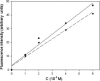
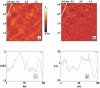
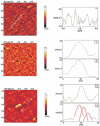
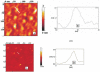
Unoxidized crystalline silicon, characterized by high purity, high homogeneity, sturdiness and an atomically flat surface, offers many advantages for the construction of electronic miniaturized biosensor arrays upon attachment of biomolecules (DNA, proteins or small organic compounds). This allows to study the incidence of molecular interactions through the simultaneous analysis, within a single experiment, of a number of samples containing small quantities of potential targets, in the presence of thousands of variables. A simple, accurate and robust methodology was established and is here presented, for the assembling of DNA sensors on the unoxidized, crystalline Si(100) surface, by loading controlled amounts of a monolayer DNA-probe through a two-step procedure. At first a monolayer of a spacer molecule, such as 10-undecynoic acid, was deposited, under optimized conditions, via controlled cathodic electrografting, then a synthetic DNA-probe was anchored to it, through amidation in aqueous solution. The surface coverage of several DNA-probes and the control of their efficiency in recognizing a complementary target-DNA upon hybridization were evaluated by fluorescence measurements. The whole process was also monitored in parallel by Atomic Force Microscopy (AFM).
Figures
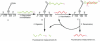


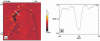

References
-
- Ball P. Silicon still supreme. Nature Mater. 2005;4:119. - PubMed
-
- Hersam M.C., Guisinger N.P., Lydin J.W. Silicon-based molecular nanotechnology. Nanotechnology. 2000;11:70–76.
-
- Yin H.B., Brown T., Greef R., Wilkinson J.S., J.Melvin J. Chemical modification and micropatterning of Si(100) with oligonucleotides. Microelectronic Eng. 2004;73–74:830–836.
-
- Herne T.M., Tarlov M.J. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 1997;119:8916–8920.
-
- Peterlinz K.A., R.M.Georgiadis R.M. Observation of hybridisation and dehybridisation of thiol-tethered DNA using two-color surface plasmon resonance spectroscopy. J. Am. Chem. Soc. 1997;119:3401–3402.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

