Inflammatory disease and lymphomagenesis caused by deletion of the Myc antagonist Mnt in T cells
- PMID: 16507988
- PMCID: PMC1430277
- DOI: 10.1128/MCB.26.6.2080-2092.2006
Inflammatory disease and lymphomagenesis caused by deletion of the Myc antagonist Mnt in T cells
Abstract
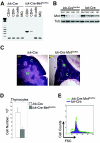
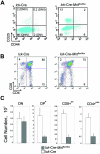
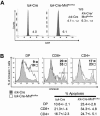
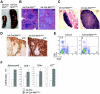
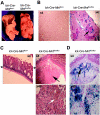
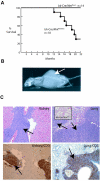
Mnt is a Max-interacting protein that can antagonize the activities of Myc oncoproteins in cultured cells. Mnt null mice die soon after birth, but conditional deletion of Mnt in breast epithelium leads to tumor formation. These and related data suggest that Mnt functions as a tumor suppressor. Here we show that conditional deletion of Mnt in T cells leads to tumor formation but also causes inflammatory disease. Deletion of Mnt caused increased apoptosis of thymic T cells and interfered with T-cell development yet led to spleen, liver, and lymph node enlargement. The proportion of T cells in the spleen and lymph nodes was reduced, and the numbers of cells in non-T-cell immune cell populations were elevated. The disruption of immune homeostasis is linked to a strong skewing toward production of T-helper 1 (Th1) cytokines and enhanced proliferation of activated Mnt-deficient CD4+ T cells. Consistent with Th1 polarization in vivo, extensive intestinal inflammation and liver necrosis developed. Finally, most mice lacking Mnt in T cells ultimately succumbed to T-cell lymphoma. These results strengthen the argument that Mnt functions as a tumor suppressor and reveal a critical and surprising role for Mnt in the regulation of T-cell development and in T-cell-dependent immune homeostasis.
Figures


Similar articles
-
Mnt-deficient mammary glands exhibit impaired involution and tumors with characteristics of myc overexpression.Cancer Res. 2006 Jun 1;66(11):5565-73. doi: 10.1158/0008-5472.CAN-05-2683. Cancer Res. 2006. PMID: 16740691
-
Of Myc and Mnt.J Cell Sci. 2006 Jan 15;119(Pt 2):208-16. doi: 10.1242/jcs.02815. J Cell Sci. 2006. PMID: 16410546 Review.
-
Mnt transcriptional repressor is functionally regulated during cell cycle progression.Oncogene. 2005 Dec 15;24(56):8326-37. doi: 10.1038/sj.onc.1208961. Oncogene. 2005. PMID: 16103876
-
Loss of the Max-interacting protein Mnt in mice results in decreased viability, defective embryonic growth and craniofacial defects: relevance to Miller-Dieker syndrome.Hum Mol Genet. 2004 May 15;13(10):1057-67. doi: 10.1093/hmg/ddh116. Epub 2004 Mar 17. Hum Mol Genet. 2004. PMID: 15028671
-
Mnt takes control as key regulator of the myc/max/mxd network.Adv Cancer Res. 2007;97:61-80. doi: 10.1016/S0065-230X(06)97003-1. Adv Cancer Res. 2007. PMID: 17419941 Review.
Cited by
-
MYC needs MNT.Cell Cycle. 2013 Feb 1;12(3):385-6. doi: 10.4161/cc.23551. Epub 2013 Jan 16. Cell Cycle. 2013. PMID: 23324353 Free PMC article. No abstract available.
-
MNT and Emerging Concepts of MNT-MYC Antagonism.Genes (Basel). 2017 Feb 20;8(2):83. doi: 10.3390/genes8020083. Genes (Basel). 2017. PMID: 28230739 Free PMC article.
-
The Multiple Faces of MNT and Its Role as a MYC Modulator.Cancers (Basel). 2021 Sep 18;13(18):4682. doi: 10.3390/cancers13184682. Cancers (Basel). 2021. PMID: 34572909 Free PMC article. Review.
-
A novel role of MNT as a negative regulator of REL and the NF-κB pathway.Oncogenesis. 2021 Jan 8;10(1):5. doi: 10.1038/s41389-020-00298-4. Oncogenesis. 2021. PMID: 33419981 Free PMC article.
-
A critical role for Mnt in Myc-driven T-cell proliferation and oncogenesis.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19685-90. doi: 10.1073/pnas.1206406109. Epub 2012 Nov 12. Proc Natl Acad Sci U S A. 2012. PMID: 23150551 Free PMC article.
References
-
- Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 6:635-645. - PubMed
-
- Blyth, K., A. Terry, M. O'Hara, E. W. Baxter, M. Campbell, M. Stewart, L. A. Donehower, D. E. Onions, J. C. Neil, and E. R. Cameron. 1995. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: contrasting effects of homozygous and heterozygous p53 loss. Oncogene 10:1717-1723. - PubMed
-
- Broussard-Diehl, C., S. R. Bauer, and R. H. Scheuermann. 1996. A role for c-myc in the regulation of thymocyte differentiation and possibly positive selection. J. Immunol. 156:3141-3150. - PubMed
-
- Buckley, A. F., C. T. Kuo, and J. M. Leiden. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-myc-dependent pathway. Nat. Immunol. 2:698-704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
