CARM1 regulates proliferation of PC12 cells by methylating HuD
- PMID: 16508003
- PMCID: PMC1430293
- DOI: 10.1128/MCB.26.6.2273-2285.2006
CARM1 regulates proliferation of PC12 cells by methylating HuD
Abstract
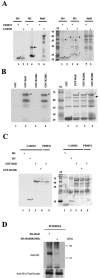
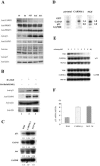
HuD is an RNA-binding protein that has been shown to induce neuronal differentiation by stabilizing labile mRNAs carrying AU-rich instability elements. Here, we show a novel mechanism of arginine methylation of HuD by coactivator-associated arginine methyltransferase 1 (CARM1) that affected mRNA turnover of p21cip1/waf1 mRNA in PC12 cells. CARM1 specifically methylated HuD in vitro and in vivo and colocalized with HuD in the cytoplasm. Inhibition of HuD methylation by CARM1 knockdown elongated the p21cip1/waf1 mRNA half-life and resulted in a slow growth rate and robust neuritogenesis in response to nerve growth factor (NGF). Methylation-resistant HuD bound more p21cip1/waf1 mRNA than did the wild type, and its overexpression upregulated p21cip1/waf1 protein expression. These results suggested that CARM1-methylated HuD maintains PC12 cells in the proliferative state by committing p21cip1/waf1 mRNA to its decay system. Since the methylated population of HuD was reduced in NGF-treated PC12 cells, downregulation of HuD methylation is a possible pathway through which NGF induces differentiation of PC12 cells.
Figures





Similar articles
-
HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects.Hum Mol Genet. 2011 Feb 1;20(3):553-79. doi: 10.1093/hmg/ddq500. Epub 2010 Nov 18. Hum Mol Genet. 2011. PMID: 21088113
-
Protein kinase C stimulates HuD-mediated mRNA stability and protein expression of neurotrophic factors and enhances dendritic maturation of hippocampal neurons in culture.Hippocampus. 2012 Dec;22(12):2303-19. doi: 10.1002/hipo.22048. Epub 2012 Jun 27. Hippocampus. 2012. PMID: 22736542
-
Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase.J Biol Chem. 2002 Nov 22;277(47):44623-30. doi: 10.1074/jbc.M206187200. Epub 2002 Sep 16. J Biol Chem. 2002. PMID: 12237300
-
CARM1/PRMT4: Making Its Mark beyond Its Function as a Transcriptional Coactivator.Trends Cell Biol. 2021 May;31(5):402-417. doi: 10.1016/j.tcb.2020.12.010. Epub 2021 Jan 20. Trends Cell Biol. 2021. PMID: 33485722 Review.
-
Role of HuD in nervous system function and pathology.Front Biosci (Schol Ed). 2013 Jan 1;5(2):554-63. doi: 10.2741/s389. Front Biosci (Schol Ed). 2013. PMID: 23277068 Review.
Cited by
-
Inhibition of coactivator-associated arginine methyltransferase 1 modulates dendritic arborization and spine maturation of cultured hippocampal neurons.J Biol Chem. 2017 Apr 14;292(15):6402-6413. doi: 10.1074/jbc.M117.775619. Epub 2017 Mar 6. J Biol Chem. 2017. PMID: 28264928 Free PMC article.
-
Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency.Nucleic Acids Res. 2017 Sep 19;45(16):9348-9360. doi: 10.1093/nar/gkx550. Nucleic Acids Res. 2017. PMID: 28637181 Free PMC article.
-
Coactivator-Associated Arginine Methyltransferase-1 Function in Alveolar Epithelial Senescence and Elastase-Induced Emphysema Susceptibility.Am J Respir Cell Mol Biol. 2015 Dec;53(6):769-81. doi: 10.1165/rcmb.2014-0216OC. Am J Respir Cell Mol Biol. 2015. PMID: 25906418 Free PMC article.
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System.Mol Neurobiol. 2023 Oct;60(10):6060-6091. doi: 10.1007/s12035-023-03465-x. Epub 2023 Jul 6. Mol Neurobiol. 2023. PMID: 37415067 Review.
-
Characterization of 3'-untranslated region of the mouse GDNF gene.BMC Mol Biol. 2012 Jan 17;13:2. doi: 10.1186/1471-2199-13-2. BMC Mol Biol. 2012. PMID: 22248285 Free PMC article.
References
-
- Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015-13021. - PubMed
-
- Akamatsu, W., H. Fujihara, T. Mitsuhashi, M. Yano, S. Shibata, Y. Hayakawa, H. J. Okano, S. Sakakibara, H. Takano, T. Takano, T. Takahashi, T. Noda, and H. Okano. 2005. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. USA 102:4625-4630. - PMC - PubMed
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. - PubMed
-
- Anderson, K. D., J. Sengupta, M. Morin, R. L. Neve, C. F. Valenzuela, and N. I. Perrone-Bizzozero. 2001. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 168:250-258. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
