Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development
- PMID: 16508010
- PMCID: PMC1430305
- DOI: 10.1128/MCB.26.6.2347-2359.2006
Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development
Abstract
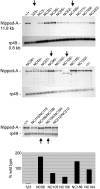
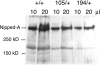
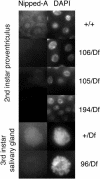
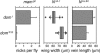
The Notch receptor controls development by activating transcription of specific target genes in response to extracellular signals. The factors that control assembly of the Notch activator complex on target genes and its ability to activate transcription are not fully known. Here we show, through genetic and molecular analysis, that the Drosophila Nipped-A protein is required for activity of Notch and its coactivator protein, mastermind, during wing development. Nipped-A and mastermind also colocalize extensively on salivary gland polytene chromosomes, and reducing Nipped-A activity decreases mastermind binding. Nipped-A is the fly homologue of the yeast Tra1 and human TRRAP proteins and is a key component of both the SAGA and Tip60 (NuA4) chromatin-modifying complexes. We find that, like Nipped-A, the Ada2b component of SAGA and the domino subunit of Tip60 are also required for mastermind function during wing development. Based on these results, we propose that Nipped-A, through the action of the SAGA and Tip60 complexes, facilitates assembly of the Notch activator complex and target gene transcription.
Figures





References
-
- Berset, T., E. F. Hoier, G. Battu, S. Canevascini, and A. Hajnal. 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291:1055-1058. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
