Structure of a putative trans-editing enzyme for prolyl-tRNA synthetase from Aeropyrum pernix K1 at 1.7 A resolution
- PMID: 16508081
- PMCID: PMC1952386
- DOI: 10.1107/S1744309104032555
Structure of a putative trans-editing enzyme for prolyl-tRNA synthetase from Aeropyrum pernix K1 at 1.7 A resolution
Abstract
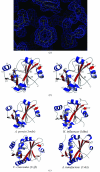
The crystal structure of APE2540, the putative trans-editing enzyme ProX from Aeropyrum pernix K1, was determined in a high-throughput manner. The crystal belongs to the monoclinic space group P2(1), with unit-cell parameters a = 47.4, b = 58.9, c = 53.6 A, beta = 106.8 degrees. The structure was solved by the multiwavelength anomalous dispersion method at 1.7 A and refined to an R factor of 16.8% (Rfree = 20.5%). The crystal structure includes two protein molecules in the asymmetric unit. Each monomer consists of eight beta-strands and seven alpha-helices. A structure-homology search revealed similarity between the trans-editing enzyme YbaK (or cysteinyl-tRNAPro deacylase) from Haemophilus influenzae (HI1434; 22% sequence identity) and putative ProX proteins from Caulobacter crescentus (16%) and Agrobacterium tumefaciens (21%).
Figures

Similar articles
-
Crystallization and preliminary crystallographic studies of putative threonyl-tRNA synthetases from Aeropyrum pernix and Sulfolobus tokodaii.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Oct 1;64(Pt 10):903-10. doi: 10.1107/S1744309108026924. Epub 2008 Sep 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18931432 Free PMC article.
-
Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.Proteins. 2000 Jul 1;40(1):86-97. doi: 10.1002/(sici)1097-0134(20000701)40:1<86::aid-prot100>3.0.co;2-y. Proteins. 2000. PMID: 10813833
-
Catalytic properties and crystal structure of thermostable NAD(P)H-dependent carbonyl reductase from the hyperthermophilic archaeon Aeropyrum pernix K1.Enzyme Microb Technol. 2016 Sep;91:17-25. doi: 10.1016/j.enzmictec.2016.05.008. Epub 2016 May 20. Enzyme Microb Technol. 2016. PMID: 27444325
-
Cloning, expression, purification, crystallization and preliminary X-ray crystallographic analyses of threonyl-tRNA synthetase editing domain from Aeropyrum pernix.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Nov 1;68(Pt 11):1390-3. doi: 10.1107/S1744309112042066. Epub 2012 Oct 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 23143256 Free PMC article.
-
Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain.J Mol Biol. 2009 Nov 27;394(2):286-96. doi: 10.1016/j.jmb.2009.09.018. Epub 2009 Sep 15. J Mol Biol. 2009. PMID: 19761773
Cited by
-
Aminoacyl-tRNA substrate and enzyme backbone atoms contribute to translational quality control by YbaK.J Phys Chem B. 2013 Apr 25;117(16):4521-7. doi: 10.1021/jp308628y. Epub 2012 Dec 6. J Phys Chem B. 2013. PMID: 23185990 Free PMC article.
-
Exclusive use of trans-editing domains prevents proline mistranslation.J Biol Chem. 2013 May 17;288(20):14391-14399. doi: 10.1074/jbc.M113.467795. Epub 2013 Apr 5. J Biol Chem. 2013. PMID: 23564458 Free PMC article.
-
Substrate-mediated fidelity mechanism ensures accurate decoding of proline codons.J Biol Chem. 2011 Sep 9;286(36):31810-20. doi: 10.1074/jbc.M111.232611. Epub 2011 Jul 18. J Biol Chem. 2011. PMID: 21768119 Free PMC article.
References
-
- An, S. & Musier-Forsyth, K. (2004). J. Biol. Chem.279, 42359–42362. - PubMed
-
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

