Crystallization and preliminary X-ray studies on the reaction center-light-harvesting 1 core complex from Rhodopseudomonas viridis
- PMID: 16508098
- PMCID: PMC1952401
- DOI: 10.1107/S1744309104028945
Crystallization and preliminary X-ray studies on the reaction center-light-harvesting 1 core complex from Rhodopseudomonas viridis
Abstract
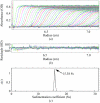
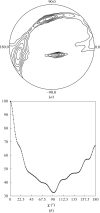
The reaction center-light-harvesting 1 (RC-LH1) core complex is the photosynthetic apparatus in the membrane of the purple photosynthetic bacterium Rhodopseudomonas viridis. The RC is surrounded by an LH1 complex that is constituted of oligomers of three types of apoproteins (alpha, beta and gamma chains) with associated bacteriochlorophyll bs and carotenoid. It has been crystallized by the sitting-drop vapour-diffusion method. A promising crystal diffracted to beyond 8.0 A resolution. It belonged to space group P1, with unit-cell parameters a = 141.4, b = 136.9, c = 185.3 A, alpha = 104.6, beta = 94.0, gamma = 110.7 degrees. A Patterson function calculated using data between 15.0 and 8.0 A resolution suggested that the LH1 complex is distributed with quasi-16-fold rotational symmetry around the RC.
Figures

Similar articles
-
Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris.Science. 2003 Dec 12;302(5652):1969-72. doi: 10.1126/science.1088892. Science. 2003. PMID: 14671305
-
Purification, characterization and crystallization of the core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum.Biochim Biophys Acta. 2007 Aug;1767(8):1057-63. doi: 10.1016/j.bbabio.2007.06.002. Epub 2007 Jun 16. Biochim Biophys Acta. 2007. PMID: 17658456
-
Characterization of a highly purified, fully active, crystallizable RC-LH1-PufX core complex from Rhodobacter sphaeroides.Photosynth Res. 2005 Nov;86(1-2):61-70. doi: 10.1007/s11120-005-5106-z. Photosynth Res. 2005. PMID: 16172926
-
A comparative look at structural variation among RC-LH1 'Core' complexes present in anoxygenic phototrophic bacteria.Photosynth Res. 2020 Aug;145(2):83-96. doi: 10.1007/s11120-020-00758-3. Epub 2020 May 19. Photosynth Res. 2020. PMID: 32430765 Free PMC article. Review.
-
Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis.EMBO J. 1989 Aug;8(8):2149-70. doi: 10.1002/j.1460-2075.1989.tb08338.x. EMBO J. 1989. PMID: 2676514 Free PMC article. Review.
Cited by
-
Various salts employed as precipitant in combination with polyethylene glycol in protein/detergent particle association.Heliyon. 2018 Dec 27;4(12):e01073. doi: 10.1016/j.heliyon.2018.e01073. eCollection 2018 Dec. Heliyon. 2018. PMID: 30603706 Free PMC article.
References
-
- Brunisholz, R. A., Jay, F., Suter, F. & Zuber, H. (1985). Biol. Chem. Hoppe-Seyler, 366, 87–98. - PubMed
-
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. - PubMed
-
- Ducruix, A. & Giegé, R. (1992). Crystallization of Nucleic Acids and Proteins. A Practical Approach, edited by A. Ducruix & R. Giegé, pp. 73–98. Oxford: IRL Press.
-
- Fotiadis, D., Qian, P., Philippsen, A., Bullough, P. A., Engel, A. & Hunter, C. N. (2004). J. Biol. Chem.279, 2063–2068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

