Nuclease sensitive element binding protein 1 associates with the selenocysteine insertion sequence and functions in mammalian selenoprotein translation
- PMID: 16508950
- PMCID: PMC3730826
- DOI: 10.1002/jcp.20619
Nuclease sensitive element binding protein 1 associates with the selenocysteine insertion sequence and functions in mammalian selenoprotein translation
Abstract
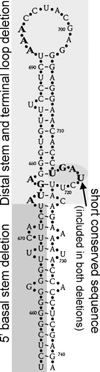
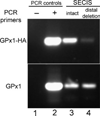
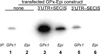
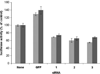
Biosynthesis of selenium-containing proteins requires insertion of the unusual amino acid selenocysteine by alternative translation of a UGA codon, which ordinarily serves as a stop codon. In eukaryotes, selenoprotein translation depends upon one or more selenocysteine insertion sequence (SECIS) elements located in the 3'-untranslated region of the mRNA, as well as several SECIS-binding proteins. Our laboratory has previously identified nuclease sensitive element binding protein 1 (NSEP1) as another SECIS-binding protein, but evidence has been presented both for and against its role in SECIS binding in vivo and in selenoprotein translation. Our current studies sought to resolve this controversy, first by investigating whether NSEP1 interacts closely with SECIS elements within intact cells. After reversible in vivo cross-linking and ribonucleoprotein immunoprecipitation, mRNAs encoding two glutathione peroxidase family members co-precipitated with NSEP1 in both human and rat cell lines. Co-immunoprecipitation of an epitope-tagged GPX1 construct depended upon an intact SECIS element in its 3'-untranslated region. To test the functional importance of this interaction on selenoprotein translation, we used small inhibitory RNAs to reduce the NSEP1 content of tissue culture cells and then examined the effect of that reduction on the activity of a SECIS-dependent luciferase reporter gene for which expression depends upon readthrough of a UGA codon. Co-transfection of small inhibitory RNAs directed against NSEP1 decreased its expression by approximately 50% and significantly reduced luciferase activity. These studies demonstrate that NSEP1 is an authentic SECIS binding protein that is structurally associated with the selenoprotein translation complex and functionally involved in the translation of selenoproteins in mammalian cells.
Copyright 2006 Wiley-Liss, Inc.
Figures



Similar articles
-
An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine.Mol Cell Biol. 1997 Apr;17(4):1977-85. doi: 10.1128/MCB.17.4.1977. Mol Cell Biol. 1997. PMID: 9121445 Free PMC article.
-
A protein binds the selenocysteine insertion element in the 3'-UTR of mammalian selenoprotein mRNAs.Nucleic Acids Res. 1996 Feb 1;24(3):464-9. doi: 10.1093/nar/24.3.464. Nucleic Acids Res. 1996. PMID: 8602359 Free PMC article.
-
Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2.RNA Biol. 2014;11(11):1402-13. doi: 10.1080/15476286.2014.996472. RNA Biol. 2014. PMID: 25692238 Free PMC article.
-
Solution structure of SECIS, the mRNA element required for eukaryotic selenocysteine insertion--interaction studies with the SECIS-binding protein SBP.Biomed Environ Sci. 1997 Sep;10(2-3):177-81. Biomed Environ Sci. 1997. PMID: 9315308 Review.
-
Post-transcriptional control of selenoprotein biosynthesis.Curr Protein Pept Sci. 2012 Jun;13(4):337-46. doi: 10.2174/138920312801619448. Curr Protein Pept Sci. 2012. PMID: 22708491 Review.
Cited by
-
Secisbp2 is essential for embryonic development and enhances selenoprotein expression.Antioxid Redox Signal. 2014 Aug 20;21(6):835-49. doi: 10.1089/ars.2013.5358. Epub 2014 Feb 4. Antioxid Redox Signal. 2014. PMID: 24274065 Free PMC article.
-
The molecular biology of selenocysteine.Biomol Concepts. 2013 Aug;4(4):349-65. doi: 10.1515/bmc-2013-0007. Biomol Concepts. 2013. PMID: 25436585 Free PMC article. Review.
-
SECIS-binding protein 2 interacts with the SMN complex and the methylosome for selenoprotein mRNP assembly and translation.Nucleic Acids Res. 2017 May 19;45(9):5399-5413. doi: 10.1093/nar/gkx031. Nucleic Acids Res. 2017. PMID: 28115638 Free PMC article.
-
Regulation and function of selenoproteins in human disease.Biochem J. 2009 Jul 29;422(1):11-22. doi: 10.1042/BJ20090219. Biochem J. 2009. PMID: 19627257 Free PMC article. Review.
-
Regulation of the extracellular antioxidant selenoprotein plasma glutathione peroxidase (GPx-3) in mammalian cells.Mol Cell Biochem. 2009 Jul;327(1-2):111-26. doi: 10.1007/s11010-009-0049-x. Epub 2009 Feb 15. Mol Cell Biochem. 2009. PMID: 19219623 Free PMC article.
References
-
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991a;353:273–276. - PubMed
-
- Berry MJ, Banu L, Larsen PR. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991b;349:438–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

