A multivariate prediction model for microarray cross-hybridization
- PMID: 16509965
- PMCID: PMC1409802
- DOI: 10.1186/1471-2105-7-101
A multivariate prediction model for microarray cross-hybridization
Abstract
Background: Expression microarray analysis is one of the most popular molecular diagnostic techniques in the post-genomic era. However, this technique faces the fundamental problem of potential cross-hybridization. This is a pervasive problem for both oligonucleotide and cDNA microarrays; it is considered particularly problematic for the latter. No comprehensive multivariate predictive modeling has been performed to understand how multiple variables contribute to (cross-) hybridization.
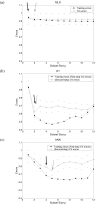
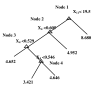
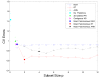
Results: We propose a systematic search strategy using multiple multivariate models [multiple linear regressions, regression trees, and artificial neural network analyses (ANNs)] to select an effective set of predictors for hybridization. We validate this approach on a set of DNA microarrays with cytochrome p450 family genes. The performance of our multiple multivariate models is compared with that of a recently proposed third-order polynomial regression method that uses percent identity as the sole predictor. All multivariate models agree that the 'most contiguous base pairs between probe and target sequences,' rather than percent identity, is the best univariate predictor. The predictive power is improved by inclusion of additional nonlinear effects, in particular target GC content, when regression trees or ANNs are used.
Conclusion: A systematic multivariate approach is provided to assess the importance of multiple sequence features for hybridization and of relationships among these features. This approach can easily be applied to larger datasets. This will allow future developments of generalized hybridization models that will be able to correct for false-positive cross-hybridization signals in expression experiments.
Figures


References
-
- Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SP. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–467. - PubMed
-
- Ptijssen P. Laboratory Techniques in Biochemistry and molecular biology: hybridization with nucleic acid probes Part I: theory and nucleic acid preparation. Vol. 24. Amsterdam, The Netherlands , Elsevier Science Publishers BV; 1993. Overview of principles of hybridization and the strategy of nucleic acid probe assays; pp. 19–78.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

