Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae
- PMID: 16510898
- PMCID: PMC1415214
- DOI: 10.1101/gr.4355406
Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae
Abstract
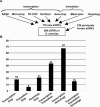
Genes with small open reading frames (sORFs; <100 amino acids) represent an untapped source of important biology. sORFs largely escaped analysis because they were difficult to predict computationally and less likely to be targeted by genetic screens. Thus, the substantial number of sORFs and their potential importance have only recently become clear. To investigate sORF function, we undertook the first functional studies of sORFs in any system, using the model eukaryote Saccharomyces cerevisiae. Based on independent experimental approaches and computational analyses, evidence exists for 299 sORFs in the S. cerevisiae genome, representing approximately 5% of the annotated ORFs. We determined that a similar percentage of sORFs are annotated in other eukaryotes, including humans, and 184 of the S. cerevisiae sORFs exhibit similarity with ORFs in other organisms. To investigate sORF function, we constructed a collection of gene-deletion mutants of 140 newly identified sORFs, each of which contains a strain-specific "molecular barcode," bringing the total number of sORF deletion strains to 247. Phenotypic analyses of the new gene-deletion strains identified 22 sORFs required for haploid growth, growth at high temperature, growth in the presence of a nonfermentable carbon source, or growth in the presence of DNA damage and replication-arrest agents. We provide a collection of sORF deletion strains that can be integrated into the existing deletion collection as a resource for the yeast community for elucidating gene function. Moreover, our analyses of the S. cerevisiae sORFs establish that sORFs are conserved across eukaryotes and have important biological functions.
Figures



Comment in
-
Small open reading frames: not so small anymore.Genome Res. 2006 Mar;16(3):314-5. doi: 10.1101/gr.4976706. Genome Res. 2006. PMID: 16510897 No abstract available.
References
-
- Adams, A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Aouida, M., Tounekti, O., Leduc, A., Belhadj, O., Mir, L., and Ramotar, D. 2004. Isolation and characterization of Saccharomyces cerevisiae mutants with enhanced resistance to the anticancer drug bleomycin. Curr. Genet. 45 265–272. - PubMed
-
- Basrai, M.A. and Hieter, P. 2002. Transcriptome analysis of Saccharomyces cerevisiae using serial analysis of gene expression. Methods Enzymol. 350 414–444. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
