Systematic identification of functional orthologs based on protein network comparison
- PMID: 16510899
- PMCID: PMC1415213
- DOI: 10.1101/gr.4526006
Systematic identification of functional orthologs based on protein network comparison
Abstract
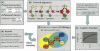
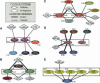
Annotating protein function across species is an important task that is often complicated by the presence of large paralogous gene families. Here, we report a novel strategy for identifying functionally related proteins that supplements sequence-based comparisons with information on conserved protein-protein interactions. First, the protein interaction networks of two species are aligned by assigning proteins to sequence homology clusters using the Inparanoid algorithm. Next, probabilistic inference is performed on the aligned networks to identify pairs of proteins, one from each species, that are likely to retain the same function based on conservation of their interacting partners. Applying this method to Drosophila melanogaster and Saccharomyces cerevisiae, we analyze 121 cases for which functional orthology assignment is ambiguous when sequence similarity is used alone. In 61 of these cases, the network supports a different protein pair than that favored by sequence comparisons. These results suggest that network analysis can be used to provide a key source of information for refining sequence-based homology searches.
Figures


References
-
- Aebersold, R. and Mann, M. 2003. Mass spectrometry-based proteomics. Nature 422: 198–207. - PubMed
-
- Aitchison, J.D., Blobel, G., and Rout, M.P. 1996. Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274: 624–627. - PubMed
-
- Besag, J. 1974. Spatial interaction and the statistical analysis of lattice systems. J. Roy. Statist. Soc. B: 192–236.
-
- Brenner, S.E. 1999. Errors in genome annotation. Trends Genet. 15: 132–133. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
