SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces
- PMID: 16512892
- PMCID: PMC1459204
- DOI: 10.1186/1471-2105-7-104
SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces
Abstract
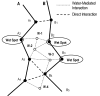
Background: Currently there is a strong need for methods that help to obtain an accurate description of protein interfaces in order to be able to understand the principles that govern molecular recognition and protein function. Many of the recent efforts to computationally identify and characterize protein networks extract protein interaction information at atomic resolution from the PDB. However, they pay none or little attention to small protein ligands and solvent. They are key components and mediators of protein interactions and fundamental for a complete description of protein interfaces. Interactome profiling requires the development of computational tools to extract and analyze protein-protein, protein-ligand and detailed solvent interaction information from the PDB in an automatic and comparative fashion. Adding this information to the existing one on protein-protein interactions will allow us to better understand protein interaction networks and protein function.
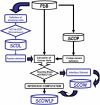
Description: SCOWLP (Structural Characterization Of Water, Ligands and Proteins) is a user-friendly and publicly accessible web-based relational database for detailed characterization and visualization of the PDB protein interfaces. The SCOWLP database includes proteins, peptidic-ligands and interface water molecules as descriptors of protein interfaces. It contains currently 74,907 protein interfaces and 2,093,976 residue-residue interactions formed by 60,664 structural units (protein domains and peptidic-ligands) and their interacting solvent. The SCOWLP web-server allows detailed structural analysis and comparisons of protein interfaces at atomic level by text query of PDB codes and/or by navigating a SCOP-based tree. It includes a visualization tool to interactively display the interfaces and label interacting residues and interface solvent by atomic physicochemical properties. SCOWLP is automatically updated with every SCOP release.
Conclusion: SCOWLP enriches substantially the description of protein interfaces by adding detailed interface information of peptidic-ligands and solvent to the existing protein-protein interaction databases. SCOWLP may be of interest to many structural bioinformaticians. It provides a platform for automatic global mapping of protein interfaces at atomic level, representing a useful tool for classification of protein interfaces, protein binding comparative studies, reconstruction of protein complexes and understanding protein networks. The web-server with the database and its additional summary tables used for our analysis are available at http://www.scowlp.org.
Figures


References
-
- Argos P. An investigation of protein subunit and domain interfaces. Protein Eng. 1988;2:101–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

