HTLV-I antisense transcripts initiating in the 3'LTR are alternatively spliced and polyadenylated
- PMID: 16512901
- PMCID: PMC1459196
- DOI: 10.1186/1742-4690-3-15
HTLV-I antisense transcripts initiating in the 3'LTR are alternatively spliced and polyadenylated
Abstract
Background: Antisense transcription in retroviruses has been suggested for both HIV-1 and HTLV-I, although the existence and coding potential of these transcripts remain controversial. Thorough characterization is required to demonstrate the existence of these transcripts and gain insight into their role in retrovirus biology.
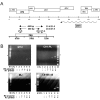
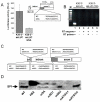
Results: This report provides the first complete characterization of an antisense retroviral transcript that encodes the previously described HTLV-I HBZ protein. In this study, we show that HBZ-encoding transcripts initiate in the 3' long terminal repeat (LTR) at several positions and consist of two alternatively spliced variants (SP1 and SP2). Expression of the most abundant HBZ spliced variant (SP1) could be detected in different HTLV-I-infected cell lines and importantly in cellular clones isolated from HTLV-I-infected patients. Polyadenylation of HBZ RNA occurred at a distance of 1450 nucleotides downstream of the HBZ stop codon in close proximity of a typical polyA signal. We have also determined that translation mostly initiates from the first exon located in the 3' LTR and that the HBZ isoform produced from the SP1 spliced variant demonstrated inhibition of Tax and c-Jun-dependent transcriptional activation.
Conclusion: These results conclusively demonstrate the existence of antisense transcription in retroviruses, which likely plays a role in HTLV-I-associated pathogenesis through HBZ protein synthesis.
Figures


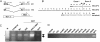


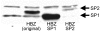
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

