A dehydration-inducible gene in the truffle Tuber borchii identifies a novel group of dehydrins
- PMID: 16512918
- PMCID: PMC1550403
- DOI: 10.1186/1471-2164-7-39
A dehydration-inducible gene in the truffle Tuber borchii identifies a novel group of dehydrins
Abstract
Background: The expressed sequence tag M6G10 was originally isolated from a screening for differentially expressed transcripts during the reproductive stage of the white truffle Tuber borchii. mRNA levels for M6G10 increased dramatically during fruiting body maturation compared to the vegetative mycelial stage.
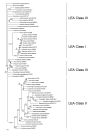
Results: Bioinformatics tools, phylogenetic analysis and expression studies were used to support the hypothesis that this sequence, named TbDHN1, is the first dehydrin (DHN)-like coding gene isolated in fungi. Homologs of this gene, all defined as "coding for hypothetical proteins" in public databases, were exclusively found in ascomycetous fungi and in plants. Although complete (or almost complete) fungal genomes and EST collections of some Basidiomycota and Glomeromycota are already available, DHN-like proteins appear to be represented only in Ascomycota. A new and previously uncharacterized conserved signature pattern was identified and proposed to Uniprot database as the main distinguishing feature of this new group of DHNs. Expression studies provide experimental evidence of a transcript induction of TbDHN1 during cellular dehydration.
Conclusion: Expression pattern and sequence similarities to known plant DHNs indicate that TbDHN1 is the first characterized DHN-like protein in fungi. The high similarity of TbDHN1 with homolog coding sequences implies the existence of a novel fungal/plant group of LEA Class II proteins characterized by a previously undescribed signature pattern.
Figures
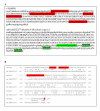


 indicates the transcription initiator. Stop codons are marked with an asterisk. AA amino acids that form the CSP; AA amino acids that form the repeated block of 46 amino acids.
indicates the transcription initiator. Stop codons are marked with an asterisk. AA amino acids that form the CSP; AA amino acids that form the repeated block of 46 amino acids.
References
-
- Campbell SA, Close TJ. Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytologist. 1997;137:61–74. doi: 10.1046/j.1469-8137.1997.00831.x. - DOI
-
- Close TJ. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiologia Plantarum. 1996;97:795–803. doi: 10.1034/j.1399-3054.1996.970422.x. - DOI
-
- Close TJ. Dehydrins: A commonality in the response of plants to dehydration and low temperature. Physiologia Plantarum. 1997;100:291–296. doi: 10.1034/j.1399-3054.1997.1000210.x. - DOI
-
- Dure LIII. Structural motifs in LEA proteins. In: Close TJ and Bray E, editor. Plant Responses to Cellular Dehydration during Environmental Stress Current Topics in Plant Physiology. Rockville, MD, American Society of Plant Physiologists; 1993. pp. 91–103.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

