A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo
- PMID: 16513737
- PMCID: PMC1428139
- DOI: 10.1128/JB.188.6.2073-2080.2006
A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo
Abstract
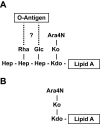
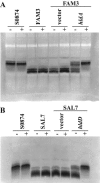
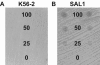
Burkholderia cenocepacia is an important opportunistic pathogen of patients with cystic fibrosis. This bacterium is inherently resistant to a wide range of antimicrobial agents, including high concentrations of antimicrobial peptides. We hypothesized that the lipopolysaccharide (LPS) of B. cenocepacia is important for both virulence and resistance to antimicrobial peptides. We identified hldA and hldD genes in B. cenocepacia strain K56-2. These two genes encode enzymes involved in the modification of heptose sugars prior to their incorporation into the LPS core oligosaccharide. We constructed a mutant, SAL1, which was defective in expression of both hldA and hldD, and by performing complementation studies we confirmed that the functions encoded by both of these B. cenocepacia genes were needed for synthesis of a complete LPS core oligosaccharide. The LPS produced by SAL1 consisted of a short lipid A-core oligosaccharide and was devoid of O antigen. SAL1 was sensitive to the antimicrobial peptides polymyxin B, melittin, and human neutrophil peptide 1. In contrast, another B. cenocepacia mutant strain that produced complete lipid A-core oligosaccharide but lacked polymeric O antigen was not sensitive to polymyxin B or melittin. As determined by the rat agar bead model of lung infection, the SAL1 mutant had a survival defect in vivo since it could not be recovered from the lungs of infected rats 14 days postinfection. Together, these data show that the B. cenocepacia LPS inner core oligosaccharide is needed for in vitro resistance to three structurally unrelated antimicrobial peptides and for in vivo survival in a rat model of chronic lung infection.
Figures

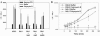

References
-
- Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

