The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20
- PMID: 16513750
- PMCID: PMC1428151
- DOI: 10.1128/JB.188.6.2207-2213.2006
The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20
Abstract
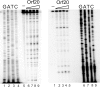
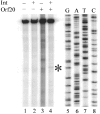
Orf20 of the conjugative transposon Tn916 was purified as a chimeric protein fused to maltose binding protein (MBP-Orf20). The chimeric protein possessed endonucleolytic activity, cleaving both strands of the Tn916 origin of conjugal transfer (oriT) at several distinct sites and favoring GT dinucleotides. Incubation of the oriT DNA with purified Tn916 integrase (Int) and MBP-Orf20 resulted in strand- and sequence-specific cleavage of oriT at a TGGT motif in the transferred strand. This motif lies immediately adjacent to a sequence in oriT previously shown to be protected from DNase I cleavage by Int. The endonucleolytic cleavages produced by Orf20 generated a 3' OH group that could be radiolabeled by dideoxy ATP and terminal transferase. The production of a 3' OH group distinguished these Orf20-dependent cleavage events from those catalyzed by Int at the ends of Tn916. Thus, Orf20 functions as the relaxase of Tn916, nicking oriT as the first step in conjugal DNA transfer. Remarkably for a tyrosine recombinase, Tn916 Int acts as a specificity factor in the reaction, conferring both strand and sequence specificities on the endonucleolytic cleavage activity of Orf20.
Figures




References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. - PubMed
-
- Caperon, M. G., and J. R. Scott. 1989. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 59:1027-1034. - PubMed
-
- Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

