Theoretical analysis of single-molecule force spectroscopy experiments: heterogeneity of chemical bonds
- PMID: 16513778
- PMCID: PMC1459525
- DOI: 10.1529/biophysj.105.077099
Theoretical analysis of single-molecule force spectroscopy experiments: heterogeneity of chemical bonds
Abstract
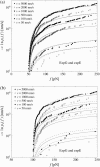
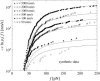
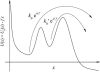
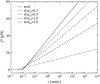
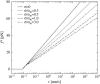
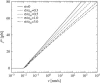
We show that the standard theoretical framework in single-molecule force spectroscopy has to be extended to consistently describe the experimental findings. The basic amendment is to take into account heterogeneity of the chemical bonds via random variations of the force-dependent dissociation rates. This results in a very good agreement between theory and rupture data from several different experiments.
Figures
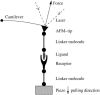
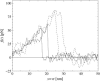

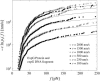
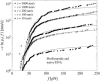




References
-
- Merkel, R. 2001. Force spectroscopy on single passive biomolecules and single biomolecular bonds. Phys. Rep. 346:343–385.
-
- Evans, E. 2001. Probing the relation between force, lifetime, and chemistry in single biomolecular bonds. Annu. Rev. Biomol. Struct. 30:105–128. - PubMed
-
- Florin, E.-L., V. T. Moy, and H. E. Gaub. 1994. Adhesion forces between individual ligand-receptor pairs. Science. 264:415–417. - PubMed
-
- Lee, G., L. A. Chrisey, and R. J. Colton. 1994. Direct measurement of the interaction forces between complementary strands of DNA with atomic force microscopy. Science. 266:771–775. - PubMed
-
- Dammer, U., O. Popescu, P. Wagner, D. Anselmetti, H.-J. Güntherodt, and G. N. Misevic. 1995. Binding strength between cell adhesion proteoglycans measured by atomic force microscopy. Science. 267:1173–1175. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

