DNA-damage response pathways triggered by viral replication
- PMID: 16515730
- PMCID: PMC4221734
- DOI: 10.1017/S1462399406010544
DNA-damage response pathways triggered by viral replication
Abstract
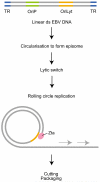
Many viruses, with distinct replication strategies, activate DNA-damage response pathways, including the lentivirus human immunodeficiency virus (HIV) and the DNA viruses Epstein-Barr virus (EBV), herpes simplex virus 1, adenovirus and SV40. DNA-damage response pathways involving DNA-dependent protein kinase, ataxia-telengiectasia mutated (ATM) and 'ataxia-telengiectasia and Rad3-related' (ATR) have all been implicated. This review focuses on the effects of HIV and EBV replication on DNA repair pathways. It has been suggested that activation of cellular DNA repair and recombination enzymes is beneficial for viral replication, as illustrated by the ability of suppressors of the ATM and ATR family to inhibit HIV replication. However, activation of DNA-damage response pathways can also promote apoptosis. Viruses can tailor the cellular response by suppressing downstream signalling from DNA-damage sensors, as exemplified by EBV. New small-molecule inhibitors of the DNA-damage response pathways could therefore be of value to treat viral infections.
Figures

Similar articles
-
Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection.J Virol. 2007 Feb;81(4):1934-50. doi: 10.1128/JVI.01670-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151099 Free PMC article.
-
Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment.J Biol Chem. 2005 Mar 4;280(9):8156-63. doi: 10.1074/jbc.M411405200. Epub 2004 Dec 15. J Biol Chem. 2005. PMID: 15611093
-
DNA Damage Signaling Is Induced in the Absence of Epstein-Barr Virus (EBV) Lytic DNA Replication and in Response to Expression of ZEBRA.PLoS One. 2015 May 7;10(5):e0126088. doi: 10.1371/journal.pone.0126088. eCollection 2015. PLoS One. 2015. PMID: 25950714 Free PMC article.
-
Epstein-Barr Virus Hijacks DNA Damage Response Transducers to Orchestrate Its Life Cycle.Viruses. 2017 Nov 16;9(11):341. doi: 10.3390/v9110341. Viruses. 2017. PMID: 29144413 Free PMC article. Review.
-
Epstein-Barr Virus (EBV)-Related Lymphoproliferative Disorders in Ataxia Telangiectasia: Does ATM Regulate EBV Life Cycle?Front Immunol. 2019 Jan 4;9:3060. doi: 10.3389/fimmu.2018.03060. eCollection 2018. Front Immunol. 2019. PMID: 30662441 Free PMC article. Review.
Cited by
-
LEDGF (p75) promotes DNA-end resection and homologous recombination.Nat Struct Mol Biol. 2012 Aug;19(8):803-10. doi: 10.1038/nsmb.2314. Epub 2012 Jul 8. Nat Struct Mol Biol. 2012. PMID: 22773103
-
Induction of DNA Damages upon Marek's Disease Virus Infection: Implication in Viral Replication and Pathogenesis.J Virol. 2017 Nov 30;91(24):e01658-17. doi: 10.1128/JVI.01658-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978699 Free PMC article.
-
Combination Therapy with Reovirus and ATM Inhibitor Enhances Cell Death and Virus Replication in Canine Melanoma.Mol Ther Oncolytics. 2019 Aug 28;15:49-59. doi: 10.1016/j.omto.2019.08.003. eCollection 2019 Dec 20. Mol Ther Oncolytics. 2019. PMID: 31650025 Free PMC article.
-
Simian virus 40 large T antigen induces IFN-stimulated genes through ATR kinase.J Immunol. 2014 Jun 15;192(12):5933-42. doi: 10.4049/jimmunol.1303470. Epub 2014 May 5. J Immunol. 2014. PMID: 24799566 Free PMC article.
-
Parvovirus B19 nonstructural protein-induced damage of cellular DNA and resultant apoptosis.Int J Med Sci. 2011 Jan 15;8(2):88-96. doi: 10.7150/ijms.8.88. Int J Med Sci. 2011. PMID: 21278893 Free PMC article.
References
-
- Shirata N, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280:30336–30341. - PubMed
-
- Weitzman MD, et al. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 2004;3:1165–1173. - PubMed
-
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. - PubMed
-
- Daniel R, Katz RA, Skalka AM. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. - PubMed
-
- Kudoh A, et al. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem. 2005;280:8156–8163. - PubMed
Further reading, resources and contacts
-
-
Review of ATM: Pandita TK. A multifaceted role for ATM in genome maintenance. Expert Rev Mol Med. 2003:1–21. 2003. PubMed: 14987398.
-
-
-
Review of HIV replication: Freed EO. HIV-1 replication. Somat Cell Mol Genet. 2001;26:13–33. PubMed: 12465460.
-
-
-
Review of EBV replication: Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev Med Virol. 2005;15:3–15. PubMed: 15386591.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

