Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus
- PMID: 16517636
- PMCID: PMC1393175
- DOI: 10.1128/AEM.72.3.1891-1899.2006
Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus
Abstract
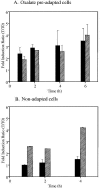
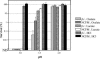
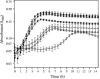
Oxalic acid is found in dietary sources (such as coffee, tea, and chocolate) or is produced by the intestinal microflora from metabolic precursors, like ascorbic acid. In the human intestine, oxalate may combine with calcium, sodium, magnesium, or potassium to form less soluble salts, which can cause pathological disorders such as hyperoxaluria, urolithiasis, and renal failure in humans. In this study, an operon containing genes homologous to a formyl coenzyme A transferase gene (frc) and an oxalyl coenzyme A decarboxylase gene (oxc) was identified in the genome of the probiotic bacterium Lactobacillus acidophilus. Physiological analysis of a mutant harboring a deleted version of the frc gene confirmed that frc expression specifically improves survival in the presence of oxalic acid at pH 3.5 compared with the survival of the wild-type strain. Moreover, the frc mutant was unable to degrade oxalate. These genes, which have not previously been described in lactobacilli, appear to be responsible for oxalate degradation in this organism. Transcriptional analysis using cDNA microarrays and reverse transcription-quantitative PCR revealed that mildly acidic conditions were a prerequisite for frc and oxc transcription. As a consequence, oxalate-dependent induction of these genes occurred only in cells first adapted to subinhibitory concentrations of oxalate and then exposed to pH 5.5. Where genome information was available, other lactic acid bacteria were screened for frc and oxc genes. With the exception of Lactobacillus gasseri and Bifidobacterium lactis, none of the other strains harbored genes for oxalate utilization.
Figures



References
-
- Abe, K., Z.-S. Ruan, and P. C. Maloney. 1996. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J. Biol. Chem. 271:6789-6793. - PubMed
-
- Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. - PMC - PubMed
-
- Andresson, H., S. Filipsson, and L. Hulten. 1978. Urinary oxalate excretion related to ileocolic surgery in patients with Crohn's disease. Scand. J. Gastroenterol. 13:465-469. - PubMed
-
- Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. - PubMed
-
- Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, R. Cano, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

