Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM
- PMID: 16517637
- PMCID: PMC1393231
- DOI: 10.1128/AEM.72.3.1900-1909.2006
Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM
Abstract
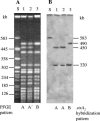
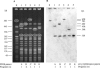
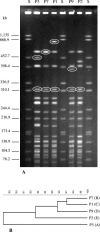
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:NM (nonmotile) is a unique clone that causes outbreaks of hemorrhagic colitis and hemolytic-uremic syndrome. In well-defined clusters of cases, we have observed significant variability in pulsed-field gel electrophoresis (PFGE) patterns which could indicate coinfection by different strains. An analysis of randomly selected progeny colonies of an outbreak strain after subcultivation demonstrated that they displayed either the cognate PFGE outbreak pattern or one of four additional patterns and were <89% similar. These profound alterations were associated with changes in the genomic position of one of two Shiga toxin 2-encoding genes (stx2) in the outbreak strain or with the loss of this gene. The two stx2 alleles in the outbreak strain were identical but were flanked with phage-related sequences with only 77% sequence identity. Neither of these phages produced plaques, but one lysogenized E. coli K-12 and integrated in yecE in the lysogens and the wild-type strain. The presence of two stx2 genes which correlated with increased production of Stx2 in vitro but not with the clinical outcome of infection was also found in 14 (21%) of 67 SF EHEC O157:NM isolates from sporadic cases of human disease. The variability of PFGE patterns for the progeny of a single colony must be considered when interpreting PFGE patterns in SF EHEC O157-associated outbreaks.
Figures



References
-
- Ammon, A., L. R. Peterson, and H. Karch. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of E. coli O157:H−. J. Infect. Dis. 179:1274-1277. - PubMed
-
- Bettelheim, K. A., M. Whipp, S. P. Djordjevic, and V. Ramachandran. 2002. First isolation outside Europe of sorbitol-fermenting verocytotoxigenic Escherichia coli (VTEC) belonging to O group O157. J. Med. Microbiol. 51:713-714. - PubMed
-
- Bielaszewska, M., H. Schmidt, A. Liesegang, R. Prager, W. Rabsch, H. Tschäpe, A. Cizek, J. Janda, K. Blahova, and H. Karch. 2000. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J. Clin. Microbiol. 38:3470-3473. - PMC - PubMed
-
- Bielaszewska, M., and H. Karch. 2005. Consequences of enterohaemorrhagic Escherichia coli infection for the vascular endothelium. Thromb. Haemost. 94:312-318. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

