Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255
- PMID: 16517654
- PMCID: PMC1393235
- DOI: 10.1128/AEM.72.3.2050-2063.2006
Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255
Abstract
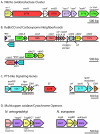
The alphaproteobacterium Nitrobacter winogradskyi (ATCC 25391) is a gram-negative facultative chemolithoautotroph capable of extracting energy from the oxidation of nitrite to nitrate. Sequencing and analysis of its genome revealed a single circular chromosome of 3,402,093 bp encoding 3,143 predicted proteins. There were extensive similarities to genes in two alphaproteobacteria, Bradyrhizobium japonicum USDA110 (1,300 genes) and Rhodopseudomonas palustris CGA009 CG (815 genes). Genes encoding pathways for known modes of chemolithotrophic and chemoorganotrophic growth were identified. Genes encoding multiple enzymes involved in anapleurotic reactions centered on C2 to C4 metabolism, including a glyoxylate bypass, were annotated. The inability of N. winogradskyi to grow on C6 molecules is consistent with the genome sequence, which lacks genes for complete Embden-Meyerhof and Entner-Doudoroff pathways, and active uptake of sugars. Two gene copies of the nitrite oxidoreductase, type I ribulose-1,5-bisphosphate carboxylase/oxygenase, cytochrome c oxidase, and gene homologs encoding an aerobic-type carbon monoxide dehydrogenase were present. Similarity of nitrite oxidoreductases to respiratory nitrate reductases was confirmed. Approximately 10% of the N. winogradskyi genome codes for genes involved in transport and secretion, including the presence of transporters for various organic-nitrogen molecules. The N. winogradskyi genome provides new insight into the phylogenetic identity and physiological capabilities of nitrite-oxidizing bacteria. The genome will serve as a model to study the cellular and molecular processes that control nitrite oxidation and its interaction with other nitrogen-cycling processes.
Figures

References
-
- Ahlers, B., W. Konig, and E. Bock. 1990. Nitrite reductase activity in Nitrobacter vulgaris. FEMS Microbiol. Lett. 67:121-126.
-
- Aleem, M. I. H., and D. L. Sewell. 1984. Oxidoreductase systems in Nitrobacter agilis, p. 185-210. In W. R. Strohl and O. H. Tuovinen (ed.), Microbial chemoautotrophy. Ohio State University Press, Columbus, Ohio.
-
- Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. - PubMed
-
- Auran, T. B., and E. L. Schmidt. 1976. Lipids of Nitrobacter and effects of cultural conditions on fatty acid composition. Biochim. Biophys. Acta 431:390-398. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

