The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa
- PMID: 16517656
- PMCID: PMC1393232
- DOI: 10.1128/AEM.72.3.2070-2079.2006
The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa
Abstract
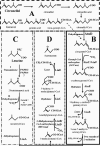
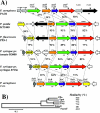
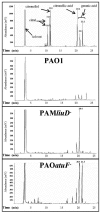
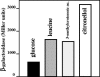
Evidence suggests that the Pseudomonas aeruginosa PAO1 gnyRDBHAL cluster, which is involved in acyclic isoprenoid degradation (A. L. Díaz-Pérez, N. A. Zavala-Hernández, C. Cervantes, and J. Campos-García, Appl. Environ. Microbiol. 70:5102-5110, 2004), corresponds to the liuRABCDE cluster (B. Hoschle, V. Gnau, and D. Jendrossek, Microbiology 151:3649-3656, 2005). A liu (leucine and isovalerate utilization) homolog cluster was found in the PAO1 genome and is related to the catabolism of acyclic monoterpenes of the citronellol family (AMTC); it was named the atu cluster (acyclic terpene utilization), consisting of the atuCDEF genes and lacking the hydroxymethyl-glutaryl-coenzyme A (CoA) lyase (HMG-CoA lyase) homolog. Mutagenesis of the atu and liu clusters showed that both are involved in AMTC and leucine catabolism by encoding the enzymes related to the geranyl-CoA and the 3-methylcrotonyl-CoA pathways, respectively. Intermediary metabolites of the acyclic monoterpene pathway, citronellic and geranic acids, were accumulated, and leucine degradation rates were affected in both atuF and liuD mutants. The alpha subunit of geranyl-CoA carboxylase and the alpha subunit of 3-methylcrotonyl-CoA carboxylase (alpha-MCCase), encoded by the atuF and liuD genes, respectively, were both induced by citronellol, whereas only the alpha-MCCase subunit was induced by leucine. Both citronellol and leucine also induced a LacZ transcriptional fusion at the liuB gene. The liuE gene encodes a probable hydroxy-acyl-CoA lyase (probably HMG-CoA lyase), an enzyme with bifunctional activity that is essential for both AMTC and leucine degradation. P. aeruginosa PAO1 products encoded by the liuABCD cluster showed a higher sequence similarity (77.2 to 79.5%) with the probable products of liu clusters from several Pseudomonas species than with the atuCDEF cluster from PAO1 (41.5%). Phylogenetic studies suggest that the atu cluster from P. aeruginosa could be the result of horizontal transfer from Alphaproteobacteria. Our results suggest that the atu and liu clusters are bifunctional operons involved in both the AMTC and leucine catabolic pathways.
Figures
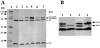

References
-
- Campos-García, J., and G. Soberón-Chávez. 2000. Degradation of the methyl substituted alkene, citronellol, by Pseudomonas aeruginosa, wild type and mutant strains. Biotechnol. Lett. 22:235-237.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

