Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms
- PMID: 16517871
- PMCID: PMC1393138
- DOI: 10.1128/JCM.44.3.881-887.2006
Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms
Abstract
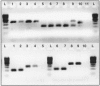
The Mycobacterium avium species consists of a group of organisms that are genetically related but phenotypically diverse, with certain variants presenting clear differences in terms of their host association and disease manifestations. The ability to distinguish between these subtypes is of relevance for accurate diagnosis and for control programs. Using a comparative genomics approach, we have uncovered large sequence polymorphisms that are, respectively, absent from bird-type M. avium isolates and from cattle types and sheep types of M. avium subsp. paratuberculosis. By evaluating the distribution of these genomic polymorphisms across a panel of strains, we were able to assign unique genomic signatures to these host-associated variants. We propose a simple PCR-based strategy based on these polymorphisms that can rapidly type M. avium isolates into these subgroups.
Figures

Similar articles
-
Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species.Mol Cell Probes. 1998 Dec;12(6):349-58. doi: 10.1006/mcpr.1998.0194. Mol Cell Probes. 1998. PMID: 9843652
-
Genomic diversity in Mycobacterium avium: single nucleotide polymorphisms between the S and C strains of M. avium subsp. paratuberculosis and with M. a. avium.Mol Cell Probes. 2007 Feb;21(1):66-75. doi: 10.1016/j.mcp.2006.08.002. Epub 2006 Aug 30. Mol Cell Probes. 2007. PMID: 17049206
-
Extensive genomic polymorphism within Mycobacterium avium.J Bacteriol. 2004 Sep;186(18):6332-4. doi: 10.1128/JB.186.18.6332-6334.2004. J Bacteriol. 2004. PMID: 15342607 Free PMC article.
-
Genetic diversity and phylogeny of Mycobacterium avium.Infect Genet Evol. 2014 Jan;21:375-83. doi: 10.1016/j.meegid.2013.12.007. Epub 2013 Dec 15. Infect Genet Evol. 2014. PMID: 24345519 Review.
-
Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions.Rev Sci Tech. 2005 Dec;24(3):1061-6. Rev Sci Tech. 2005. PMID: 16642774 Review.
Cited by
-
Genetic diversity of Mycobacterium avium subspecies paratuberculosis and the influence of strain type on infection and pathogenesis: a review.Vet Res. 2015 Jun 19;46(1):64. doi: 10.1186/s13567-015-0203-2. Vet Res. 2015. PMID: 26092160 Free PMC article. Review.
-
Comparative Genomics of Mycobacterium avium Subspecies Paratuberculosis Sheep Strains.Front Vet Sci. 2021 Feb 15;8:637637. doi: 10.3389/fvets.2021.637637. eCollection 2021. Front Vet Sci. 2021. PMID: 33659287 Free PMC article.
-
Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans.BMC Evol Biol. 2007 Sep 27;7:177. doi: 10.1186/1471-2148-7-177. BMC Evol Biol. 2007. PMID: 17900363 Free PMC article.
-
Epidemiological evidence for Mycobacterium avium subspecies paratuberculosis as a cause of Crohn's disease.Epidemiol Infect. 2007 Oct;135(7):1057-68. doi: 10.1017/S0950268807008448. Epub 2007 Apr 20. Epidemiol Infect. 2007. PMID: 17445316 Free PMC article. Review.
-
Complete Genome Sequence of Ovine Mycobacterium avium subsp. paratuberculosis Strain JIII-386 (MAP-S/type III) and Its Comparison to MAP-S/type I, MAP-C, and M. avium Complex Genomes.Microorganisms. 2020 Dec 29;9(1):70. doi: 10.3390/microorganisms9010070. Microorganisms. 2020. PMID: 33383865 Free PMC article.
References
-
- Behr, M. A., M. Semret, A. Poon, and E. Schurr. 2004. Crohn's disease, mycobacteria, and NOD2. Lancet Infect. Dis. 4:136-137. - PubMed
-
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. - PubMed
-
- Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. - PMC - PubMed
-
- Chacon, O., L. E. Bermudez, and R. G. Barletta. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329-363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

