Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002
- PMID: 16517889
- PMCID: PMC1393141
- DOI: 10.1128/JCM.44.3.999-1017.2006
Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002
Abstract
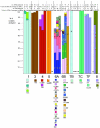
Monitoring of serotypes and their clonal associations is critical as pneumococci adapt to the selective pressures exerted by the pneumococcal seven-valent conjugate vaccine (PCV7). We genotyped 1,476 invasive isolates from the Active Bacterial Core surveillance (705 [89.8%] of the isolates were obtained from children <5 years of age, and 771 [18.4%] of the isolates were obtained from individuals >5 years of age) in 2001 and 2002 (after the introduction of PCV7). The data were compared to the results for 1,168 invasive isolates (855 [83.9%] of the isolates were from children <5 years of age) collected in 1999. Among children <5 years of age, the incidence of invasive disease due to non-PCV7 serogroups together with serogroup 19A increased (P < 0.001). Eighty-three clonal sets, representing 177 multilocus sequence types (STs), were compiled from the 3-year isolate set. Among the non-PCV7 serogroups, newly emerging clones were uncommon; and a significant expansion of already established clones occurred for serotypes 3 (ST180), 7F (ST191), 15BCF (ST199), 19A (ST199), 22F (ST433), 33F (ST662), and 38 (ST393). However, additional minor clonal types within serotypes 1, 6A, 6B, 7C, 9N, 10A, 12F, 14, 15B/C, 17F, 19A, 19F, 20, 22F, and 33F that were absent in 1999 were found during 2001 and 2002. Although 23 clonal sets exhibited multiple serotypes, for most serotypes there were either no changes or modest changes in clonal compositions since the introduction of PCV7. The only example of an identical ST shared between non-PCV7 and PCV7 or PCV7-related serotypes was ST199; however, ST199 was prevalent within serotypes 15B/C and 19A before and after PCV7 introduction. Continued genotypic surveillance is warranted, since certain clones not targeted by PCV7 are expanding, and their emergence as significant pathogens could occur with maintained vaccine pressure.
Figures




References
-
- Active Bacterial Core Surveillance. 26. December 2004, posting date. http://www.cdc.gov/ncidod/dbmd/abcs/.
-
- Beall, B., M. C. McEllistrem, R. E. Gertz, D. J. Boxrud, J. M. Besser, L. H. Harrison, J. H. Jorgensen, C. G. Whitney, and the Active Bacterial Core Surveillance/Emerging Infections Program Network. 2002. Emergence of a novel penicillin-nonsusceptible, invasive serotype 35B clone of Streptococcus pneumoniae within the United States. J. Infect. Dis. 186:118-122. - PubMed
-
- Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. - PubMed
-
- Brueggemann, A. B., T. E. Peto, D. W. Crook, J. C. Butler, K. G. Kristinsson, and B. G. Spratt. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J. Infect. Dis. 190:1203-1211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

