Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections
- PMID: 16517890
- PMCID: PMC1393109
- DOI: 10.1128/JCM.44.3.1018-1028.2006
Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections
Abstract
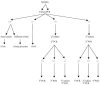
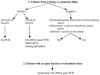
The 16S rRNA gene PCR in the diagnosis of bone and joint infections has not been systematically tested. Five hundred twenty-five bone and joint samples collected from 525 patients were cultured and submitted to 16S rRNA gene PCR detection of bacteria in parallel. The amplicons with mixed sequences were also cloned. When discordant results were observed, culture and PCR were performed once again. Bacteria were detected in 139 of 525 samples. Culture and 16S rRNA gene PCR yielded identical documentation in 475 samples. Discrepancies were linked to 13 false-positive culture results, 5 false-positive PCR results, 9 false-negative PCR results, 16 false-negative culture results, and 7 mixed infections. Cloning and sequencing of 16S rRNA gene amplicons in 6 of 8 patients with mixed infections identified 2 to 8 bacteria per sample. Rarely described human pathogens such as Alcaligenes faecalis, Comamonas terrigena, and 21 anaerobes were characterized. We also detected, by 16S rRNA gene PCR, four previously identified bacteria never reported in human infection, Alkanindiges illinoisensis, dehydroabietic acid-degrading bacterium DhA-73, unidentified Hailaer soda lake bacterium, and uncultured bacterium clone HuCa4. Seven organisms representing new potential species were also detected. PCR followed by cloning and sequencing may help to identify new pathogens involved in mixed bone infection.
Figures



References
-
- Balsam, L. B., G. M. Sheperd, and K. L. Ruoff. 1997. Streptococcus anginosus spondylodiskitis. Clin. Infect. Dis. 24:93-94. - PubMed
-
- Bartz, H., C. Nonnenmacher, C. Bollmann, M. Kuhl, S. Zimmermann, K. Heeg, and R. Mutters. 2005. Micromonas (Peptostreptococcus) micros: unusual case of prosthetic infection associated with dental procedures. Int. J. Med. Microbiol. 294:465-470. - PubMed
-
- Belzunegui, J., J. R. De Rios, J. J. Intxausti, and J. A. Iribarren. 2000. Septic arthritis caused by Stenotrophomonas maltophilia in a patient with acquired immunodeficiency syndrome. Clin. Exp. Rheumatol. 18:265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

