DNA sequences shaped by selection for stability
- PMID: 16518467
- PMCID: PMC1378130
- DOI: 10.1371/journal.pgen.0020022
DNA sequences shaped by selection for stability
Abstract
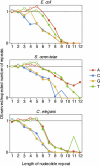
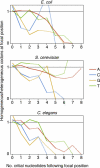
The sequence of a stretch of nucleotides affects its propensity for errors during replication and expression. Are proteins encoded by stable or unstable nucleotide sequences? If selection for variability is prevalent, one could expect an excess of unstable sequences. Alternatively, if selection against targets for errors were substantial, an excess of stable sequences would be expected. We screened the genome sequences of different organisms for an important determinant of stability, the presence of mononucleotide repeats. We find that codons are used to encode proteins in a way that avoids the emergence of mononucleotide repeats, and we can attribute this bias to selection rather than a neutral process. This indicates that selection for stability, rather than for the generation of variation, substantially influences how information is encoded in the genome.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
-
- Moxon ER, Rainey PB, Nowak MA, Lenski RE. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. - PubMed
-
- Chang DK, Metzgar D, Wills C, Boland CR. Microsatellites in the eukaryotic DNA mismatch repair genes as modulators of evolutionary mutation rate. Genome Res. 2001;11:1145–1146. - PubMed
-
- Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. - PubMed
-
- King DG, Soller M, Kashi Y. Evolutionary tuning knobs. Endeavour. 1997;21:36–40.
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

