Colocalization of somatostatin receptors and epidermal growth factor receptors in breast cancer cells
- PMID: 16519802
- PMCID: PMC1479379
- DOI: 10.1186/1475-2867-6-5
Colocalization of somatostatin receptors and epidermal growth factor receptors in breast cancer cells
Abstract
Background: Somatostatin receptor (SSTR) expression is positively correlated with tumor size and inversely correlated with epidermal growth factor receptor (ErbB) levels and tumor differentiation. In the present study, we compared SSTR1-5 and ErbB1-4 mRNA and protein expression in two breast cancer cell lines: MCF-7 (ER+) and MDA-MB-231 (ERalpha-).
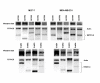
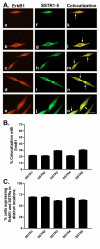
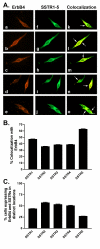
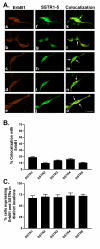
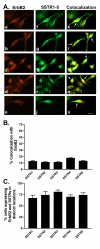
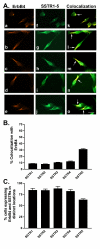
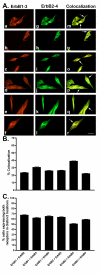
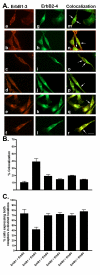
Results: All five SSTRs and four ErbBs were variably expressed as both cell surface and cytoplasmic proteins. In both cell lines, SSTR4 and SSTR1 were highly expressed, followed by SSTR2 and SSTR5 with SSTR3 being the least expressed subtype, at the protein level. ErbBs were variably expressed with ErbB1 as the predominant subtype in both cell lines. ErbB1 is followed by ErbB3, ErbB2 and ErbB4 in MCF-7 at both the protein and mRNA levels. In MDA-MB-231 cells, ErbB1 is followed by ErbB2, ErbB4 and ErbB3. Our results indicate significant correlations at the level of mRNA and protein expression in a cell and receptor-specific manner. Using indirect immunofluorescence, we found that, in MCF-7 cells, SSTR5 was the most prominent subtype coexpressed with ErbBs followed by SSTR3, SSTR4, SSTR1 and SSTR2, respectively. In MDA-MB-231 cells, SSTR1 colocalized strongly with ErbBs followed by SSTR5, SSTR4, SSTR3 and SSTR2. ErbBs displayed higher levels of colocalization amongst themselves in MCF-7 cells than in MDA-MB-231 cells.
Conclusion: These findings may explain the poor response to endocrine therapy in ER-cancer. Differential distribution of SSTR subtypes with ErbBs in breast cancer cells in a receptor-specific manner may be considered as a novel diagnosis for breast tumors.
Figures



Similar articles
-
Somatostatin receptors in primary human breast cancer: quantitative analysis of mRNA for subtypes 1--5 and correlation with receptor protein expression and tumor pathology.Breast Cancer Res Treat. 2005 Jul;92(2):175-86. doi: 10.1007/s10549-005-2414-0. Breast Cancer Res Treat. 2005. PMID: 15986128
-
Subtype-selective expression of the five somatostatin receptors (hSSTR1-5) in human pancreatic islet cells: a quantitative double-label immunohistochemical analysis.Diabetes. 1999 Jan;48(1):77-85. doi: 10.2337/diabetes.48.1.77. Diabetes. 1999. PMID: 9892225
-
Somatostatin receptors 1 and 5 heterodimerize with epidermal growth factor receptor: agonist-dependent modulation of the downstream MAPK signalling pathway in breast cancer cells.Cell Signal. 2009 Mar;21(3):428-39. doi: 10.1016/j.cellsig.2008.11.012. Epub 2008 Dec 3. Cell Signal. 2009. PMID: 19070659
-
Somatostatin receptors in pituitary and development of somatostatin receptor subtype-selective analogs.Endocrine. 2003 Apr;20(3):265-9. doi: 10.1385/ENDO:20:3:265. Endocrine. 2003. PMID: 12721506 Review.
-
Somatostatin and its receptor family.Front Neuroendocrinol. 1999 Jul;20(3):157-98. doi: 10.1006/frne.1999.0183. Front Neuroendocrinol. 1999. PMID: 10433861 Review.
Cited by
-
Ablation of neuropsin-neuregulin 1 signaling imbalances ErbB4 inhibitory networks and disrupts hippocampal gamma oscillation.Transl Psychiatry. 2017 Mar 7;7(3):e1052. doi: 10.1038/tp.2017.20. Transl Psychiatry. 2017. PMID: 28267150 Free PMC article.
-
Cross-talk and modulation of signaling between somatostatin and growth factor receptors.Endocrine. 2011 Oct;40(2):168-80. doi: 10.1007/s12020-011-9524-8. Epub 2011 Aug 26. Endocrine. 2011. PMID: 21870170 Review.
-
Somatostatin-dopamine ligands in the treatment of pituitary adenomas.Rev Endocr Metab Disord. 2009 Jun;10(2):83-90. doi: 10.1007/s11154-008-9086-0. Epub 2008 Jul 24. Rev Endocr Metab Disord. 2009. PMID: 18651224 Review.
-
The ERBB3 receptor in cancer and cancer gene therapy.Cancer Gene Ther. 2008 Jul;15(7):413-48. doi: 10.1038/cgt.2008.15. Epub 2008 Apr 11. Cancer Gene Ther. 2008. PMID: 18404164 Free PMC article. Review.
-
Expression and selective activation of somatostatin receptor subtypes induces cell cycle arrest in cancer cells.Oncol Lett. 2019 Feb;17(2):1723-1731. doi: 10.3892/ol.2018.9773. Epub 2018 Nov 28. Oncol Lett. 2019. PMID: 30675231 Free PMC article.
References
-
- Patel YC. Basic aspects of somatostatin receptors. In: LeRoith D, editor. Advances in Molecular and Cellular Endocrinology. Greenwich, CT , JAI Press; 1998.
-
- Ciocca DR, Puy LA, Fasoli LC, Tello O, Aznar JC, Gago FE, Papa SI, Sonego R. Corticotropin releasing hormone, leutinizing hormone releasing hormone, growth hormone releasing hormone, and somatostatin-like immunoreactivities in biopsies from breast cancer patients. Breast Cancer Res Treat. 1990;15:175–184. doi: 10.1007/BF01806354. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

